3BSU
 
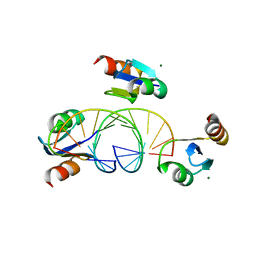 | | Hybrid-binding domain of human RNase H1 in complex with 12-mer RNA/DNA | | Descriptor: | DNA (5'-D(*DGP*DAP*DAP*DTP*DCP*DAP*DGP*DGP*(5IU)P*DGP*DTP*DC)-3'), MAGNESIUM ION, RNA (5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3'), ... | | Authors: | Nowotny, M, Cerritelli, S.M, Ghirlando, R, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2007-12-26 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD.
Embo J., 27, 2008
|
|
1ZBI
 
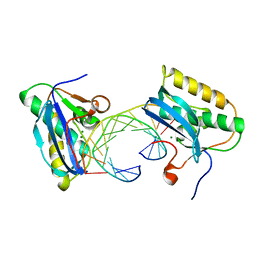 | | Bacillus halodurans RNase H catalytic domain mutant D132N in complex with 12-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3', 5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3', MAGNESIUM ION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZBF
 
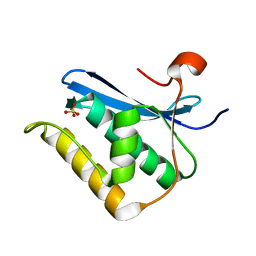 | | Crystal structure of B. halodurans RNase H catalytic domain mutant D132N | | Descriptor: | SULFATE ION, ribonuclease H-related protein | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZBL
 
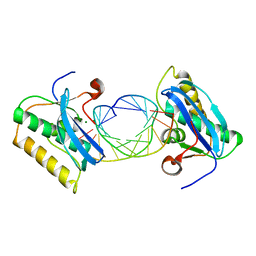 | | Bacillus halodurans RNase H catalytic domain mutant D192N in complex with 12-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3', 5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3', MAGNESIUM ION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
2QK9
 
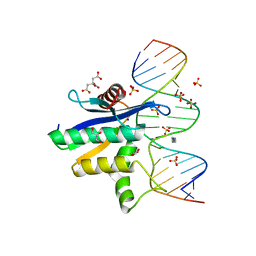 | | Human RNase H catalytic domain mutant D210N in complex with 18-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*T)-3', 5'-R(*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3', CITRATE ANION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
2QKB
 
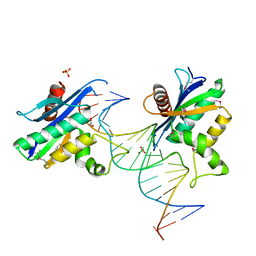 | | Human RNase H catalytic domain mutant D210N in complex with 20-mer RNA/DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*DGP*DGP*DAP*DAP*DTP*DCP*DAP*DGP*DGP*DTP*DGP*DTP*DCP*DGP*DCP*DAP*DCP*DTP*DCP*DT)-3', 5'-R(*GP*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3'), ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
2QKK
 
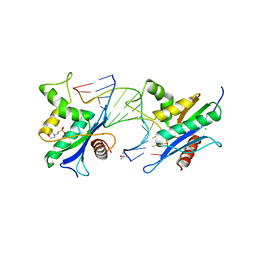 | | Human RNase H catalytic domain mutant D210N in complex with 14-mer RNA/DNA hybrid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*G)-3', ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-11 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
2G8I
 
 | |
2G8W
 
 | |
2G8V
 
 | |
2G8K
 
 | |
2G8H
 
 | |
2G8F
 
 | |
2G8U
 
 | |
7O24
 
 | |
6ZE8
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 | | Descriptor: | 5-(4-((2S,5S)-5-(4-chlorobenzyl)-2-methylmorpholino)piperidin-1-yl)-4H-1,2,4-triazol-3-amine, Chitotriosidase-1, GLYCEROL, ... | | Authors: | Nowotny, M, Bartlomiejczak, A, Napiorkowska-Gromadzka, A. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of OATD-01 , a First-in-Class Chitinase Inhibitor as Potential New Therapeutics for Idiopathic Pulmonary Fibrosis.
J.Med.Chem., 63, 2020
|
|
9I8V
 
 | | Cryo-EM structure of the Danio rerio tRNA ligase complex | | Descriptor: | ATP-dependent RNA helicase, Ashwin, Family with sequence similarity 98, ... | | Authors: | Chamera, S, Zajko, W, Czarnocki-Cieciura, M, Jaciuk, M, Koziej, L, Nowak, J, Wycisk, K, Sroka, M, Chramiec-Glabik, A, Smietanski, M, Golebiowski, F, Warminski, M, Jemielity, J, Glatt, S, Nowotny, M. | | Deposit date: | 2025-02-06 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural and biochemical characterization of the 3'-5' tRNA splicing ligases.
J.Biol.Chem., 301, 2025
|
|
4LD0
 
 | | T. thermophilus RuvC in complex with Holliday junction substrate | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, DNA 11-MER, DNA 13-MER, ... | | Authors: | Gorecka, K.M, Komorowska, W, Nowotny, M. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Crystal structure of RuvC resolvase in complex with Holliday junction substrate.
Nucleic Acids Res., 41, 2013
|
|
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
8AUP
 
 | | Structure of hARG1 with a novel inhibitor. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[(1~{R},3~{R},4~{S})-3-azanyl-3-carboxy-4-[(dimethylamino)methyl]cyclohexyl]ethyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, ... | | Authors: | Napiorkowska-Gromadzka, A, Nowak, E, Nowotny, M. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Arginase 1/2 Inhibitor OATD-02: From Discovery to First-in-man Setup in Cancer Immunotherapy.
Mol.Cancer Ther., 22, 2023
|
|
5J5Y
 
 | | Translation initiation factor 4E in complex with m2(7,2'O)GppCCl2ppG mRNA 5' cap analog | | Descriptor: | 2-amino-9-{5-O-[(R)-{[(S)-{dichloro[(R)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2-O-methyl-beta-D-ribofuranosyl}-7-methyl-9H-purin-7-ium-6-olate, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
5J5O
 
 | | Translation initiation factor 4E in complex with m7GppppG mRNA 5' cap analog | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]-7-methylguanosine, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
6F3H
 
 | | Crystal structure of Dss1 exoribonuclease active site mutant D477N from Candida glabrata | | Descriptor: | Exoribonuclease II, mitochondrial, MAGNESIUM ION, ... | | Authors: | Razew, M, Nowak, E, Nowotny, M. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components.
Nat Commun, 9, 2018
|
|
6F4A
 
 | | Yeast mitochondrial RNA degradosome complex mtEXO | | Descriptor: | Exoribonuclease II, mitochondrial, RNA (5'-R(P*AP*GP*AP*UP*AP*C)-3'), ... | | Authors: | Razew, M, Nowak, E, Nowotny, M. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components.
Nat Commun, 9, 2018
|
|
8BPR
 
 | | Complex of RecF-RecO-RecR-DNA from Thermus thermophilus (low resolution reconstruction). | | Descriptor: | DNA repair protein RecO, DNA replication and repair protein RecF, MAGNESIUM ION, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
