8IJZ
 
 | | anti-VEGF mutant | | Descriptor: | anti-VEGF nanobody | | Authors: | Qian, F, Zhu, S.Q. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
8JBR
 
 | | Structure of McyA2-CAPCP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, McyA protein | | Authors: | Peng, Y.J. | | Deposit date: | 2023-05-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modular catalytic activity of nonribosomal peptide synthetases depends on the dynamic interaction between adenylation and condensation domains.
Structure, 32, 2024
|
|
7F4A
 
 | |
8KEC
 
 | | Cyanophage A-1(L) tail fiber | | Descriptor: | Long tail fiber, short tail fiber | | Authors: | Yu, R.C, Li, Q, Zhou, C.Z. | | Deposit date: | 2023-08-11 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the intact tail machine of Anabaena myophage A-1(L).
Nat Commun, 15, 2024
|
|
7E36
 
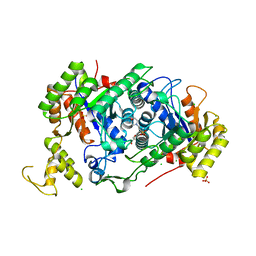 | | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkanesulfonate monooxygenase SsuD/methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase (Luciferase family), ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis.
Nat Commun, 12, 2021
|
|
7E0P
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 4-(((6-Bromo-1H-indazol-4-yl)amino)methyl)phenol (2) | | Descriptor: | 4-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]phenol, ACETIC ACID, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.635 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E5W
 
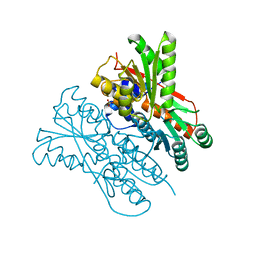 | | The structure of CcpA from Staphylococcus aureus | | Descriptor: | Catabolite control protein A, SULFATE ION | | Authors: | Yu, G, Wei, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Regulation of DNA-binding activity of the Staphylococcus aureus catabolite control protein A by copper (II)-mediated oxidation.
J.Biol.Chem., 298, 2022
|
|
7E0T
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1R,2S)-2-(((5-bromo-1H-indazol-4-yl)amino)methyl)Cyclohexan-1-ol (36) | | Descriptor: | (1~{R},2~{S})-2-[[(5-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0O
 
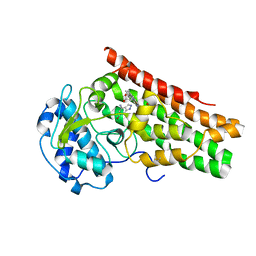 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 6-Bromo-1H-indazol-4-amine (1) | | Descriptor: | 6-bromanyl-1~{H}-indazol-4-amine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.337 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0S
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1R,2S)-2-(((6-Bromo-1H-indazol-4-yl)amino)methyl)cyclohexan-1-ol (23) | | Descriptor: | (1~{R},2~{S})-2-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, ACETIC ACID, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0U
 
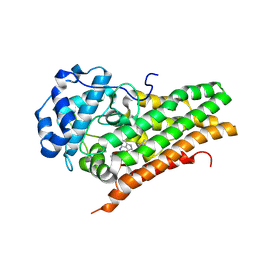 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 6-Bromo-N-(((1S,2S)-2-chlorocyclohexyl)methyl)-1H-indazol-4-amine (39) | | Descriptor: | 6-bromanyl-~{N}-[[(1~{S},2~{S})-2-chloranylcyclohexyl]methyl]-1~{H}-indazol-4-amine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0Q
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1S,2R)-2-(((6-Bromo-1H-indazol-4-yl)amino)methyl)cyclohexan-1-ol (22) | | Descriptor: | (1~{S},2~{R})-2-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7U6R
 
 | |
7URS
 
 | |
7URQ
 
 | |
7WHT
 
 | |
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7UX8
 
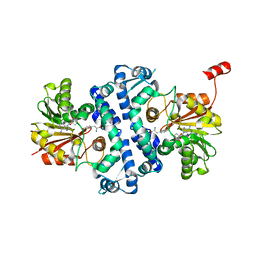 | | Crystal structure of MfnG, an L- and D-tyrosine O-methyltransferase from the marformycin biosynthesis pathway of Streptomyces drozdowiczii, with SAH and L-Tyrosine bound at 1.4 A resolution (P212121 - form II) | | Descriptor: | MfnG, S-ADENOSYL-L-HOMOCYSTEINE, TYROSINE, ... | | Authors: | Miller, M.D, Wu, K.-L, Xu, W, Xiao, H, Philips Jr, G.N. | | Deposit date: | 2022-05-05 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.
Protein Sci., 31, 2022
|
|
7UX7
 
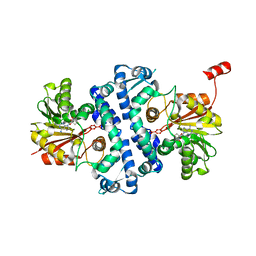 | | Crystal structure of MfnG, an L- and D-tyrosine O-methyltransferase from the marformycin biosynthesis pathway of Streptomyces drozdowiczii, with SAH bound at 1.2 A resolution (P212121 - form II) | | Descriptor: | MfnG, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Miller, M.D, Wu, K.-L, Xu, W, Xiao, H, Philips Jr, G.N. | | Deposit date: | 2022-05-05 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.
Protein Sci., 31, 2022
|
|
7UX6
 
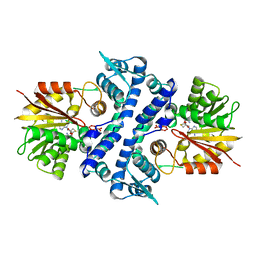 | | Crystal structure of MfnG, an L- and D-tyrosine O-methyltransferase from the marformycin biosynthesis pathway of Streptomyces drozdowiczii, with SAH bound at 1.35 A resolution (P212121 - form I) | | Descriptor: | MfnG, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Miller, M.D, Wu, K.-L, Xu, W, Xiao, H, Philips Jr, G.N. | | Deposit date: | 2022-05-05 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.
Protein Sci., 31, 2022
|
|
7WHS
 
 | |
7WHR
 
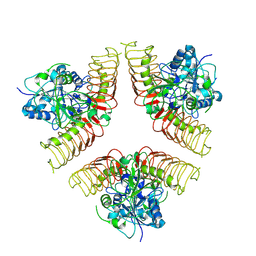 | |
7W66
 
 | | Crystal structure of a PSH1 mutant in complex with ligand | | Descriptor: | PSH1, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6C
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
