5O9Y
 
 | | Crystal structure of ScGas2 in complex with compound 11 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(3-pyridin-1-ium-1-ylpropyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5OA6
 
 | | Crystal structure of ScGas2 in complex with compound 12 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(3-quinolin-1-ium-1-ylpropyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5O9Q
 
 | | Crystal structure of ScGas2 in complex with compound 6 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(phenylmethoxymethyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,3-beta-glucanosyltransferase GAS2, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5O6C
 
 | | Crystal Structure of a threonine-selective RCR E3 ligase | | Descriptor: | E3 ubiquitin-protein ligase MYCBP2, ZINC ION | | Authors: | Pao, K.-C, Rafie, K.Z, van Aalten, D, Virdee, S. | | Deposit date: | 2017-06-06 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity.
Nature, 556, 2018
|
|
5OA2
 
 | | Crystal structure of ScGas2 in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, SULFATE ION, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5O9P
 
 | | Crystal structure of Gas2 in complex with compound 10 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(pyridin-2-ylmethoxymethyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2 | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
6TDG
 
 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 2 | | Descriptor: | 2-chloranyl-3-(4~{H}-1,2,4-triazol-3-yl)aniline, ACETYL COENZYME *A, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
1K39
 
 | | The structure of yeast delta3-delta2-enoyl-COA isomerase complexed with octanoyl-COA | | Descriptor: | OCTANOYL-COENZYME A, PHOSPHATE ION, d3,d2-enoyl CoA isomerase ECI1 | | Authors: | Mursula, A.M, Geerlof, A, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: |
|
|
5LVV
 
 | | Human OGT in complex with UDP and fused substrate peptide (Tab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit,UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Raimi, O. | | Deposit date: | 2016-09-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats.
Open Biol, 7, 2017
|
|
1H0I
 
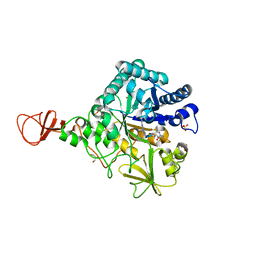 | | Complex of a chitinase with the natural product cyclopentapeptide argifin from Gliocladium | | Descriptor: | ARGIFIN, CHITINASE B, GLYCEROL, ... | | Authors: | Houston, D.R, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H0G
 
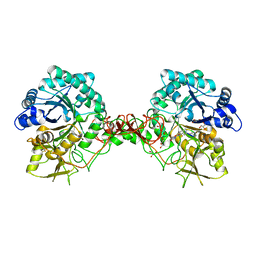 | | Complex of a chitinase with the natural product cyclopentapeptide argadin from Clonostachys | | Descriptor: | Argadin, CHITINASE B, GLYCEROL | | Authors: | Houston, D, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4BQH
 
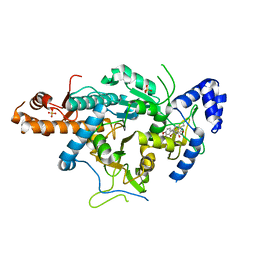 | | Crystal structure of the uridine diphosphate N-acetylglucosamine pyrophosphorylase from Trypanosoma brucei in complex with inhibitor | | Descriptor: | (3S)-3-[2-(1,3-benzodioxol-5-yl)-2-oxidanylidene-ethyl]-4-bromanyl-5-methyl-3-oxidanyl-1H-indol-2-one, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Fang, W, Raimi, O.G, vanAalten, D.M.F. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Allosteric Inhibitor of the Uridine Diphosphate N-Acetylglucosamine Pyrophosphorylase from Trypanosoma Brucei.
Acs Chem.Biol., 8, 2013
|
|
2J4O
 
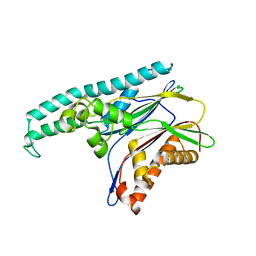 | | Structure of TAB1 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 1 | | Authors: | van Aalten, D. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tak1-Binding Protein 1 is a Pseudophosphatase.
Biochem.J., 399, 2006
|
|
2VWI
 
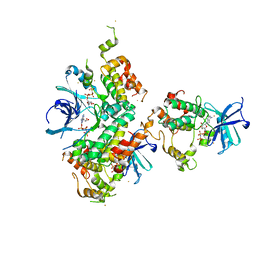 | | Structure of the OSR1 kinase, a hypertension drug target | | Descriptor: | GOLD ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Villa, F, Deak, M, Alessi, D.R, vanAalten, D.M.F. | | Deposit date: | 2008-06-25 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the Osr1 Kinase, a Hypertension Drug Target.
Proteins, 73, 2008
|
|
5OAW
 
 | |
5O9X
 
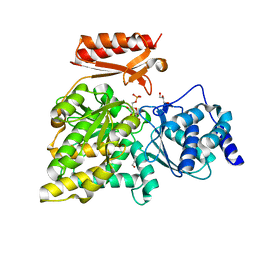 | | Crystal structure of Aspergillus fumigatus N-acetylphosphoglucosamine mutate S69A in complex with glucose1,6bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raimi, O.G, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence for substrate-assisted catalysis inN-acetylphosphoglucosamine mutase.
Biochem. J., 475, 2018
|
|
4A2Z
 
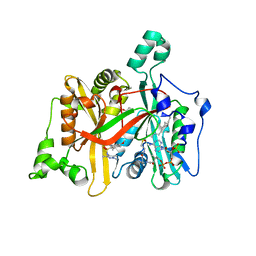 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 4-METHOXY-2,3,6-TRIMETHYL-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
2DF3
 
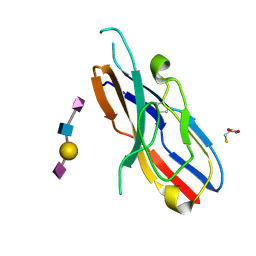 | |
1NKO
 
 | | Energetic and structural basis of sialylated oligosaccharide recognition by the natural killer cell inhibitory receptor p75/AIRM1 or Siglec-7 | | Descriptor: | Sialic acid binding Ig-like lectin 7 | | Authors: | Dimasi, N, Attril, H, van Aalten, D.M.F, Moretta, L, Biassoni, R, Mariuzza, R.A. | | Deposit date: | 2003-01-03 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2WSA
 
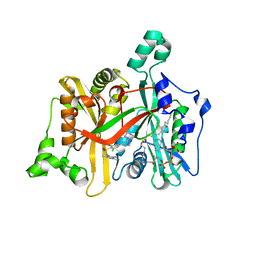 | | Crystal Structure of Leishmania major N-myristoyltransferase (NMT) with bound myristoyl-CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | N-Myristoyltransferase Inhibitors as New Leads to Treat Sleeping Sickness.
Nature, 464, 2010
|
|
1Z5M
 
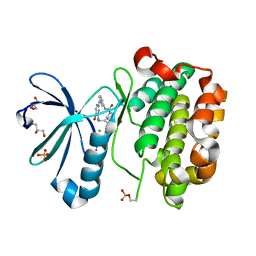 | |
2PE2
 
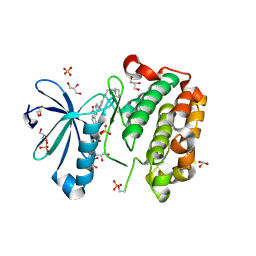 | |
2PE1
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1 (PDK1) {2-Oxo-3-[1-(1H-pyrrol-2-yl)-eth-(Z)-ylidene]-2,3-dihydro-1H-indol-5-yl}-urea {BX-517} COMPLEX | | Descriptor: | 1-{2-OXO-3-[(1R)-1-(1H-PYRROL-2-YL)ETHYL]-2H-INDOL-5-YL}UREA, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Whitlow, M, Adler, M. | | Deposit date: | 2007-04-01 | | Release date: | 2007-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Indolinone based phosphoinositide-dependent kinase-1 (PDK1) inhibitors. Part 1: Design, synthesis and biological activity.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PE0
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1 (PDK1) 5-Hydroxy-3-[1-(1H-pyrrol-2-yl)-eth-(Z)-ylidene]-1,3-dihydro-indol-2-one COMPLEX | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 5-HYDROXY-3-[(1R)-1-(1H-PYRROL-2-YL)ETHYL]-2H-INDOL-2-ONE, GLYCEROL, ... | | Authors: | Whitlow, M, Adler, M. | | Deposit date: | 2007-04-01 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Indolinone based phosphoinositide-dependent kinase-1 (PDK1) inhibitors. Part 1: Design, synthesis and biological activity.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
