7FBL
 
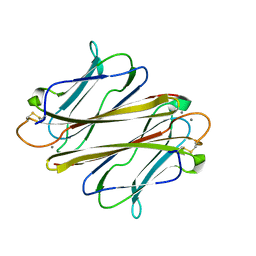 | | Thrombocorticin in complex with Ca2+ and mannose | | Descriptor: | CALCIUM ION, Thrombocorticin, alpha-D-mannopyranose | | Authors: | Kageyama, H, Onodera, K, Sakai, R, Tanaka, Y. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | A marine sponge-derived lectin reveals hidden pathway for thrombopoietin receptor activation.
Nat Commun, 13, 2022
|
|
5X52
 
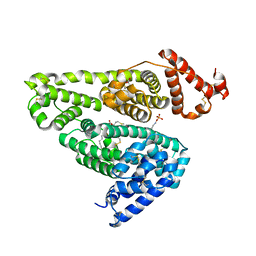 | | Human serum albumin complexed with octanoate and N-acetyl-L-methionine | | Descriptor: | N-ACETYLMETHIONINE, OCTANOIC ACID (CAPRYLIC ACID), PHOSPHATE ION, ... | | Authors: | Kawai, A, Otagiri, M. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Crystallographic analysis of the ternary complex of octanoate and N-acetyl-l-methionine with human serum albumin reveals the mode of their stabilizing interactions
Biochim. Biophys. Acta, 1865, 2017
|
|
5YO8
 
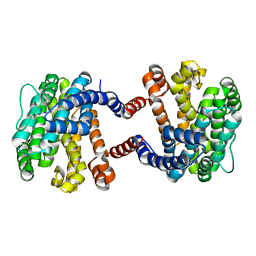 | |
1CC0
 
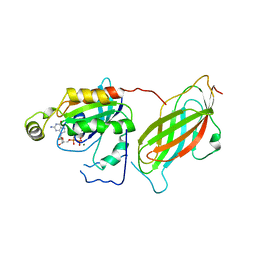 | | CRYSTAL STRUCTURE OF THE RHOA.GDP-RHOGDI COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, rho GDP dissociation inhibitor alpha, ... | | Authors: | Longenecker, K.L, Read, P, Derewenda, U, Dauter, Z, Garrard, S, Walker, L, Somlyo, A.V, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 1999-03-03 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | How RhoGDI binds Rho.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5ZC6
 
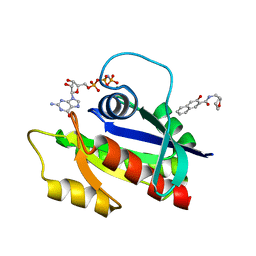 | | Solution structure of H-RasT35S mutant protein in complex with KBFM123 | | Descriptor: | 3-oxidanyl-~{N}-[[(2~{R})-oxolan-2-yl]methyl]naphthalene-2-carboxamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Matsumoto, S, Hayashi, Y, Hiraga, T, Matsuo, K, Kataoka, T. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis for Allosteric Inhibition of GTP-Bound H-Ras Protein by a Small-Molecule Compound Carrying a Naphthalene Ring
Biochemistry, 57, 2018
|
|
5B3H
 
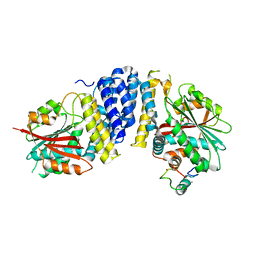 | | The crystal structure of the JACKDAW/IDD10 bound to the heterodimeric SHR-SCR complex | | Descriptor: | Protein SCARECROW, Protein SHORT-ROOT, ZINC ION, ... | | Authors: | Hirano, Y, Suyama, T, Nakagawa, M, Hakoshima, T. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
5B3G
 
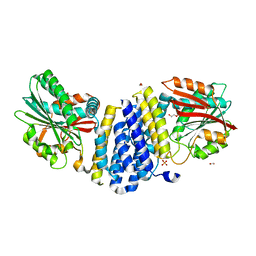 | | The crystal structure of the heterodimer of SHORT-ROOT and SCARECROW GRAS domains | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Hirano, Y, Nakagawa, M, Hakoshima, T. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
1J10
 
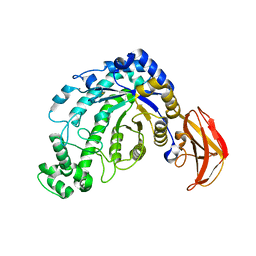 | | beta-amylase from Bacillus cereus var. mycoides in complex with GGX | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J11
 
 | | beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, Beta-amylase, CALCIUM ION | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J12
 
 | | Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG | | Descriptor: | 2-[(2S)-oxiran-2-yl]ethyl alpha-D-glucopyranoside, Beta-amylase, CALCIUM ION | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J0Z
 
 | | Beta-amylase from Bacillus cereus var. mycoides in complex with maltose | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J0Y
 
 | | Beta-amylase from Bacillus cereus var. mycoides in complex with glucose | | Descriptor: | Beta-amylase, CALCIUM ION, beta-D-glucopyranose | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
2M68
 
 | | NMR solution structure ensemble of 3-4D mutant domain 11 IGF2R in complex with IGF2 (domain 11 structure only) | | Descriptor: | Insulin-like growth factor 2 receptor variant | | Authors: | Strickland, M, Williams, C, Richards, E, Minnall, L, Crump, M.P, Frago, S, Hughes, J, Garner, L, Hoppe, H, Rezgui, D, Zaccheo, O.J, Prince, S.N, Hassan, A.B, Whittaker, S. | | Deposit date: | 2013-03-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4BIN
 
 | | Crystal structure of the E. coli N-acetylmuramoyl-L-alanine amidase AmiC | | Descriptor: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE AMIC, SODIUM ION, ZINC ION | | Authors: | Kerff, F, Rocaboy, M, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2013-04-12 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Crystal Structure of the Cell Division Amidase Amic Reveals the Fold of the Amin Domain, a New Peptidoglycan Binding Domain.
Mol.Microbiol., 90, 2013
|
|
2M6T
 
 | | NMR solution structure ensemble of 3-4D mutant domain 11 IGF2R | | Descriptor: | Insulin-like growth factor 2 receptor variant | | Authors: | Strickland, M, Williams, C, Richards, E, Minnall, L, Crump, M.P, Frago, S, Hughes, J, Garner, L, Hoppe, H, Rezgui, D, Zaccheo, O.J, Prince, S.N, Hassan, A.B, Whittaker, S. | | Deposit date: | 2013-04-09 | | Release date: | 2014-10-15 | | Last modified: | 2016-06-01 | | Method: | SOLUTION NMR | | Cite: | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4QRY
 
 | | the ground state and the N intermediate of pharaonis halorhodopsin in complex with bromide ion | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, BROMIDE ION, Halorhodopsin, ... | | Authors: | Kouyama, T, Kawaguchi, H. | | Deposit date: | 2014-07-02 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the L1, L2, N, and O States of pharaonis Halorhodopsin
Biophys.J., 108, 2015
|
|
6M5O
 
 | | Co-crystal structure of human serine hydroxymethyltransferase 2 in complex with Pyridoxal 5'-phosphate (PLP) and glycodeoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCINE, Serine hydroxymethyltransferase, ... | | Authors: | Ota, T, Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K, Sando, S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.30000663 Å) | | Cite: | Structural basis for selective inhibition of human serine hydroxymethyltransferase by secondary bile acid conjugate.
Iscience, 24, 2021
|
|
3VHZ
 
 | |
3VI0
 
 | |
8IM5
 
 | |
6M5W
 
 | | Co-crystal structure of human serine hydroxymethyltransferase 1 in complex with Pyridoxal 5'-phosphate (PLP) and glycodeoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ota, T, Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K, Sando, S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for selective inhibition of human serine hydroxymethyltransferase by secondary bile acid conjugate.
Iscience, 24, 2021
|
|
5M57
 
 | | Nek2 bound to arylaminopurine 6 | | Descriptor: | O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE, Serine/threonine-protein kinase Nek2 | | Authors: | Bayliss, R. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of purine-based probes for selective Nek2 inhibition.
Oncotarget, 8, 2017
|
|
5M51
 
 | | Nek2 bound to arylaminopurine compound 8 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-9~{H}-purin-2-yl]amino]benzamide, Serine/threonine-protein kinase Nek2 | | Authors: | Bayliss, R, Yeoh, S. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure-guided design of purine-based probes for selective Nek2 inhibition.
Oncotarget, 8, 2017
|
|
5M53
 
 | | Nek2 bound to arylaminopurine inhibitor 11 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N},~{N}-dimethyl-benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bayliss, R, Carr, K.H. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided design of purine-based probes for selective Nek2 inhibition.
Oncotarget, 8, 2017
|
|
5M55
 
 | | Nek2 bound to arylaminopurine 71 | | Descriptor: | 6-[(~{Z})-2-(diethylamino)ethenyl]-~{N}-phenyl-7~{H}-purin-2-amine, CHLORIDE ION, SULFATE ION, ... | | Authors: | Bayliss, R. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided design of purine-based probes for selective Nek2 inhibition.
Oncotarget, 8, 2017
|
|
