5TR4
 
 | | Structure of Ubiquitin activating enzyme (Uba1) in complex with ubiquitin and TAK-243 | | Descriptor: | Ubiquitin, Ubiquitin-activating enzyme E1 1, [(1~{R},2~{R},3~{S},4~{R})-2,3-bis(oxidanyl)-4-[[2-[3-(trifluoromethylsulfanyl)phenyl]pyrazolo[1,5-a]pyrimidin-7-yl]amino]cyclopentyl]methyl sulfamate | | Authors: | Sintchak, M.D. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification and Characterization of a Small Molecule Inhibitor of the Ubiquitin Activating Enzyme (TAK-243)
To be published
|
|
6LR9
 
 | | HSP90 in complex with Debio0932 | | Descriptor: | 2-[[6-(dimethylamino)-1,3-benzodioxol-5-yl]sulfanyl]-1-[2-(2,2-dimethylpropylamino)ethyl]imidazo[4,5-c]pyridin-4-amine, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Complex crystal structure determination and anti-non-small-cell lung cancer activity of the Hsp90 N inhibitor Debio0932.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
8Y0Q
 
 | | Complex of FMDV O/18074 and inter-serotype broadly neutralizing antibodies pOA-2 | | Descriptor: | VP1 of capsid protein, VP2 of capsid protein, VP3 of capsid protein, ... | | Authors: | Wu, S, Lei, D. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Discovery, recognized antigenic structures, and evolution of cross-serotype broadly neutralizing antibodies from porcine B-cell repertoires against foot-and-mouth disease virus.
Plos Pathog., 20, 2024
|
|
7T9U
 
 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
7T9V
 
 | | Crystal structure of hSTING with the agonist, SHR171032 | | Descriptor: | (3S,4S)-4-(3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazol-1-yl}propyl)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
8Y0R
 
 | | Complex of FMDV A/WH/CHA/09 and inter-serotype broadly neutralizing antibodies pOA-2 | | Descriptor: | VP1 of capsid protein, VP2 of capsid protein, VP3 of capsid protein, ... | | Authors: | Wu, S, Lei, D. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Discovery, recognized antigenic structures, and evolution of cross-serotype broadly neutralizing antibodies from porcine B-cell repertoires against foot-and-mouth disease virus.
Plos Pathog., 20, 2024
|
|
7EVN
 
 | | The cryo-EM structure of the DDX42-SF3b complex | | Descriptor: | ATP-dependent RNA helicase DDX42, PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome
Nat.Struct.Mol.Biol., 2024
|
|
7EVO
 
 | | The cryo-EM structure of the human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, RNA helicase, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
5HXG
 
 | | STM1697-FlhD complex | | Descriptor: | Flagellar transcriptional regulator FlhD, Uncharacterized protein STM1697 | | Authors: | Li, B, Yuan, Z, Qin, L, Gu, L. | | Deposit date: | 2016-01-30 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of STM1697-FlhD complex
To Be Published
|
|
5HXF
 
 | |
8HR7
 
 | | Structure of RdrA-RdrB complex | | Descriptor: | Adenosine deaminase, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRC
 
 | | Structure of dodecameric RdrB cage | | Descriptor: | Adenosine deaminase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR9
 
 | | Structure of tetradecameric RdrA ring | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR8
 
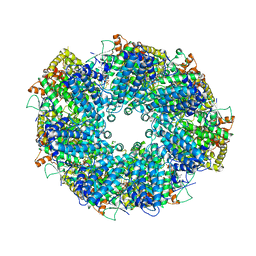 | | Structure of heptameric RdrA ring | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRB
 
 | | Structure of tetradecameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRA
 
 | | Structure of heptameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(P*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
7YDG
 
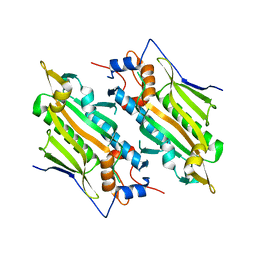 | | Crystal structure of human SARS2 catalytic domain with a disease related mutation | | Descriptor: | Serine--tRNA ligase, mitochondrial | | Authors: | Wu, S, Li, P, Zhou, X.L, Fang, P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Selective degradation of tRNASer(AGY) is the primary driver for mitochondrial seryl-tRNA synthetase-related disease.
Nucleic Acids Res., 50, 2022
|
|
