9KBT
 
 | |
9KBQ
 
 | |
9KBS
 
 | |
8Y0R
 
 | | Complex of FMDV A/WH/CHA/09 and inter-serotype broadly neutralizing antibodies pOA-2 | | Descriptor: | VP1 of capsid protein, VP2 of capsid protein, VP3 of capsid protein, ... | | Authors: | Wu, S, Lei, D. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Discovery, recognized antigenic structures, and evolution of cross-serotype broadly neutralizing antibodies from porcine B-cell repertoires against foot-and-mouth disease virus.
Plos Pathog., 20, 2024
|
|
5CZW
 
 | | Crystal structure of myroilysin | | Descriptor: | Myroilysin, ZINC ION | | Authors: | Zhou, J, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2015-08-01 | | Release date: | 2016-08-03 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Myroilysin is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Activated by a Cysteine-switch Mechanism
J. Biol. Chem., 292, 2017
|
|
2A5Y
 
 | | Structure of a CED-4/CED-9 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Apoptosis regulator ced-9, MAGNESIUM ION, ... | | Authors: | Yan, N, Liu, Q, Hao, Q, Gu, L, Shi, Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-10-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans.
Nature, 437, 2005
|
|
5GZR
 
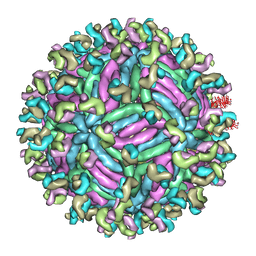 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | Descriptor: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | Authors: | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | Deposit date: | 2016-10-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6IDE
 
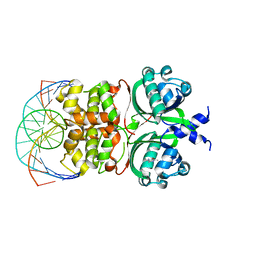 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
2LSP
 
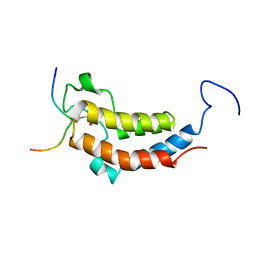 | | solution structures of BRD4 second bromodomain with NF-kB-K310ac peptide | | Descriptor: | Bromodomain-containing protein 4, NF-kB-K310ac peptide | | Authors: | Zhang, G, Liu, R, Zhong, Y, Plotnikov, A.N, Zhang, W, Rusinova, E, Gerona-Nevarro, G, Moshkina, N, Joshua, J, Chuang, P.Y, Ohlmeyer, M, He, J, Zhou, M.-M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Down-regulation of NF-kappa B transcriptional activity in HIV-associated kidney disease by BRD4 inhibition.
J.Biol.Chem., 287, 2012
|
|
3T0H
 
 | |
3T0Z
 
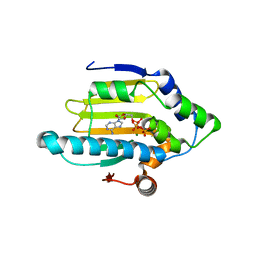 | | Hsp90 N-terminal domain bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
3T10
 
 | | HSP90 N-terminal domain bound to ACP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
5ZCX
 
 | | Structure of T20/N39 | | Descriptor: | Envelope glycoprotein, Envelope glycoprotein gp160 | | Authors: | Zhang, X.J, Ding, X.H. | | Deposit date: | 2018-02-21 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional characterization of HIV-1 cell fusion inhibitor T20.
AIDS, 33, 2019
|
|
8X43
 
 | | human KCNQ2-CaM-Ebio1-S1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(4-azanyl-1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-11-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
9J3Z
 
 | | Cryo-EM structure of lysosome cholesterol sencing protein in L state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal cholesterol signaling protein,G protein-coupled receptor 155 fusion protein | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2024-08-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for cholesterol sensing of LYCHOS and its interaction with indoxyl sulfate.
Nat Commun, 16, 2025
|
|
9J3X
 
 | | Cryo-EM structure of lysosome cholesterol sencing protein LYCHOS in Tight-state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2024-08-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for cholesterol sensing of LYCHOS and its interaction with indoxyl sulfate.
Nat Commun, 16, 2025
|
|
9J40
 
 | | Cryo-EM structure of lysosome cholesterol sencing protein complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-SULFOOXY-1H-INDOLE, ... | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2024-08-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for cholesterol sensing of LYCHOS and its interaction with indoxyl sulfate.
Nat Commun, 16, 2025
|
|
4M57
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 from maize | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10 | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
4M59
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
5MEC
 
 | |
8WR2
 
 | | Crystal Structure of Human Pyridoxal Kinase with bound Luteolin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Fan, J, Zhu, Y. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and characterization of natural product luteolin as an effective inhibitor of human pyridoxal kinase.
Bioorg.Chem., 143, 2024
|
|
5TRL
 
 | |
5MEA
 
 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 2. | | Descriptor: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5MEB
 
 | | Crystal structure of yeast Cdt1 C-terminal domain | | Descriptor: | Cell division cycle protein CDT1, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
