7NKX
 
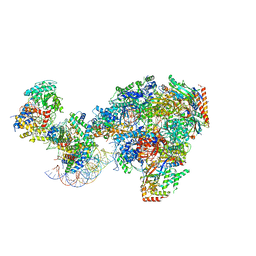 | | RNA polymerase II-Spt4/5-nucleosome-Chd1 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin elongation factor SPT4, ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OKX
 
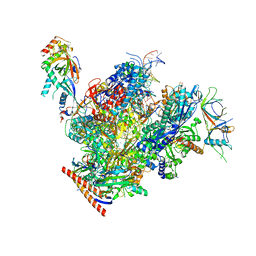 | | Structure of active transcription elongation complex Pol II-DSIF (SPT5-KOW5)-ELL2-EAF1 (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | Deposit date: | 2021-05-18 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
7OL0
 
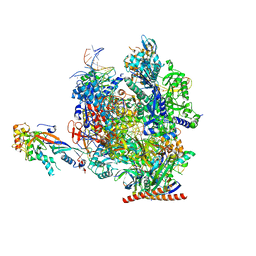 | | Structure of active transcription elongation complex Pol II-DSIF (SPT5-KOW5) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | Deposit date: | 2021-05-18 | | Release date: | 2021-07-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
7OKY
 
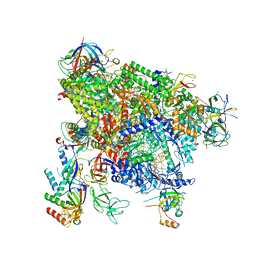 | | Structure of active transcription elongation complex Pol II-DSIF-ELL2-EAF1(composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | Deposit date: | 2021-05-18 | | Release date: | 2021-07-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
7OHA
 
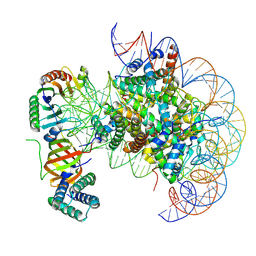 | | nucleosome with TBP and TFIIA bound at SHL +2 | | Descriptor: | DNA (122-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OH9
 
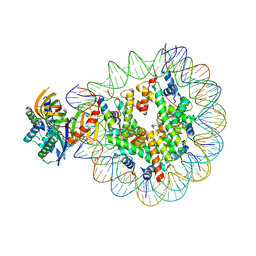 | | Nucleosome with TBP and TFIIA bound at SHL -6 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHB
 
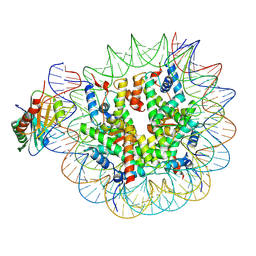 | | TBP-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-10 | | Release date: | 2021-07-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHC
 
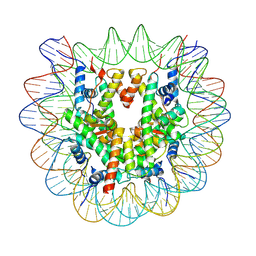 | |
7OYG
 
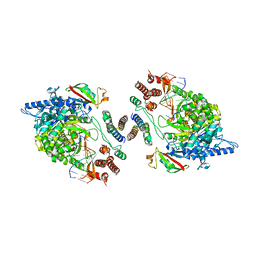 | | Dimeric form of SARS-CoV-2 RNA-dependent RNA polymerase | | Descriptor: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase (nsp12), ... | | Authors: | Jochheim, F.A, Tegunov, D, Hillen, H.S, Schmitzova, J, Kokic, G, Dienemann, C, Cramer, P. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | The structure of a dimeric form of SARS-CoV-2 polymerase
Communications Biology, 4, 2021
|
|
7OZN
 
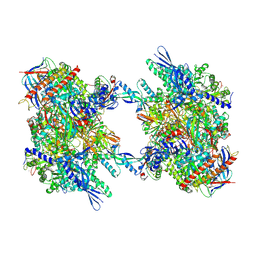 | | RNA Polymerase II dimer (Class 1) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Aibara, S, Dienemann, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of an inactive RNA polymerase II dimer.
Nucleic Acids Res., 49, 2021
|
|
7OZP
 
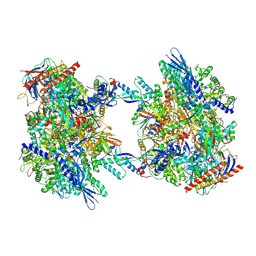 | | RNA Polymerase II dimer (Class 3) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Aibara, S, Dienemann, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of an inactive RNA polymerase II dimer.
Nucleic Acids Res., 49, 2021
|
|
7OOP
 
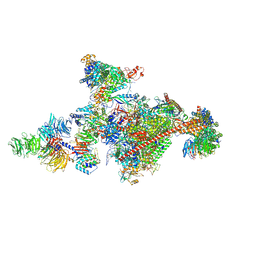 | | Pol II-CSB-CSA-DDB1-UVSSA-PAF-SPT6 (Structure 3) | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OO3
 
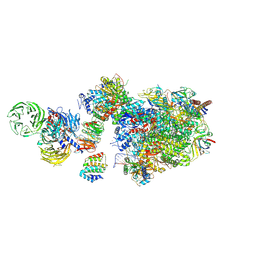 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | Descriptor: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OZO
 
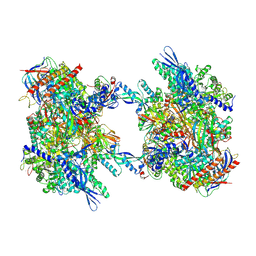 | | RNA Polymerase II dimer (Class 2) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Aibara, S, Dienemann, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of an inactive RNA polymerase II dimer.
Nucleic Acids Res., 49, 2021
|
|
7OPD
 
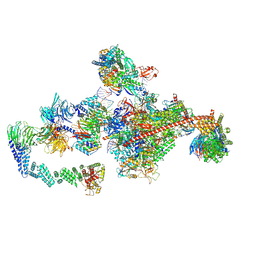 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 5) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-06 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OOB
 
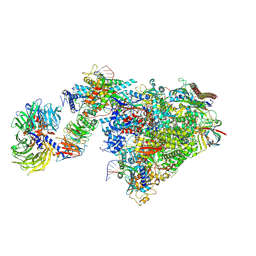 | | Pol II-CSB-CSA-DDB1-UVSSA-ADPBeF3 (Structure2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA damage-binding protein 1, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OPC
 
 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 4) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-13 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
5M3F
 
 | | Yeast RNA polymerase I elongation complex at 3.8A | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Neyer, S, Kunz, M, Geiss, C, Hantsche, M, Hodirnau, V.-V, Seybert, A, Engel, C, Scheffer, M.P, Cramer, P, Frangakis, A.S. | | Deposit date: | 2016-10-14 | | Release date: | 2016-11-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of RNA polymerase I transcribing ribosomal DNA genes.
Nature, 540, 2016
|
|
5M3M
 
 | | Free monomeric RNA polymerase I at 4.0A resolution | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Neyer, S, Kunz, M, Geiss, C, Hantsche, M, Hodirnau, V.-V, Seybert, A, Engel, C, Scheffer, M.P, Cramer, P, Frangakis, A.S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of RNA polymerase I transcribing ribosomal DNA genes.
Nature, 540, 2016
|
|
5MY1
 
 | | E. coli expressome | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kohler, R, Mooney, R.A, Mills, D.J, Kostrewa, D, Landick, R, Cramer, P. | | Deposit date: | 2017-01-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Architecture of a transcribing-translating expressome.
Science, 356, 2017
|
|
5N61
 
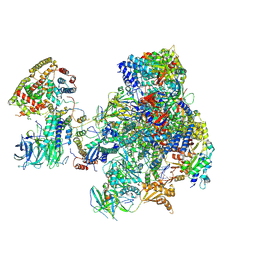 | | RNA polymerase I initially transcribing complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
6RYR
 
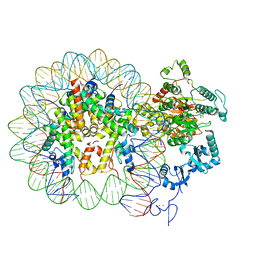 | | Nucleosome-CHD4 complex structure (single CHD4 copy) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | Authors: | Farnung, L, Ochmann, M, Cramer, P. | | Deposit date: | 2019-06-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
6RO4
 
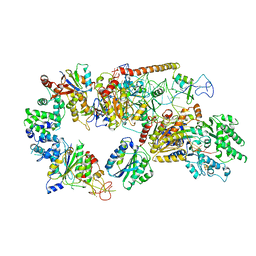 | | Structure of the core TFIIH-XPA-DNA complex | | Descriptor: | DNA repair protein complementing XP-A cells, DNA1, DNA2, ... | | Authors: | Kokic, G, Chernev, A, Tegunov, D, Dienemann, C, Urlaub, H, Cramer, P. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of TFIIH activation for nucleotide excision repair.
Nat Commun, 10, 2019
|
|
6RFL
 
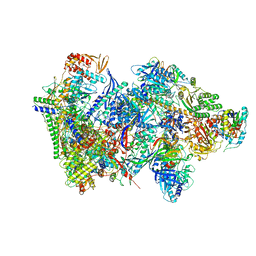 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-15 | | Release date: | 2019-12-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
6T7A
 
 | | Structure of human Sox11 transcription factor in complex with a nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Dodonova, S.O, Zhu, F, Dienemann, C, Taipale, J, Cramer, P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Nature, 580, 2020
|
|
