4U0W
 
 | |
4U0Y
 
 | |
4UAI
 
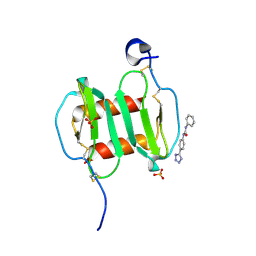 | | Crystal structure of CXCL12 in complex with inhibitor | | Descriptor: | 1-phenyl-3-[4-(1H-tetrazol-5-yl)phenyl]urea, SULFATE ION, Stromal cell-derived factor 1 | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2014-08-09 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Novel Small Molecule Ligand Bound to the CXCL12 Chemokine.
J.Med.Chem., 57, 2014
|
|
1Q4R
 
 | | Gene Product of At3g17210 from Arabidopsis Thaliana | | Descriptor: | MAGNESIUM ION, protein At3g17210 | | Authors: | Phillips Jr, G.N, Bingman, C.A, Johnson, K.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein from gene At3g17210 of Arabidopsis thaliana
Proteins, 57, 2004
|
|
3YPI
 
 | |
6G81
 
 | | Solution structure of the Ni metallochaperone HypA from Helicobacter pylori | | Descriptor: | Hydrogenase maturation factor HypA, ZINC ION | | Authors: | Spronk, C.A.E.M, Zerko, S, Gorka, M, Kozminski, W, Bardiaux, B, Zambelli, B, Musiani, F, Piccioli, M, Hu, H, Maroney, M, Ciurli, S. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of Helicobacter pylori nickel-chaperone HypA: an integrated approach using NMR spectroscopy, functional assays and computational tools.
J. Biol. Inorg. Chem., 23, 2018
|
|
6GY3
 
 | | Crystal Structure of C. glutamicum AmtR bound to glnA operator DNA | | Descriptor: | AmtR protein, DNA (5'-D(*GP*TP*CP*TP*AP*TP*AP*GP*AP*TP*CP*GP*AP*TP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*AP*TP*CP*GP*AP*TP*CP*TP*AP*TP*AP*GP*AP*C)-3') | | Authors: | Sevvana, M, Muller, Y.A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The role of DNA flexibility in transcription regulator AmtR-DNA interaction
To be published
|
|
1FDM
 
 | |
6ZTB
 
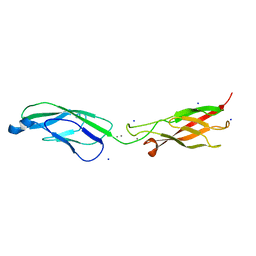 | | Crystal Structure of human P-Cadherin EC1_EC2 | | Descriptor: | CALCIUM ION, Cadherin-3, SODIUM ION | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTF
 
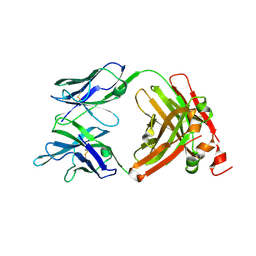 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 | | Descriptor: | CQY684 Fab heavy-chain, CQY684 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTR
 
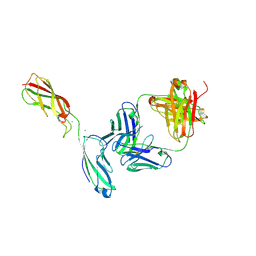 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 in complex with human P-Cadherin(108-324) | | Descriptor: | CALCIUM ION, CQY684 Fab heavy-chain, CQY684 Fab light-chain, ... | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
1G7F
 
 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177496 | | Descriptor: | 2-{4-[(2S)-2-[({[(1S)-1-CARBOXY-2-PHENYLETHYL]AMINO}CARBONYL)AMINO]-3-OXO-3-(PENTYLAMINO)PROPYL]PHENOXY}MALONIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Bleasdale, J.E, Ogg, D, Larsen, S.D. | | Deposit date: | 2000-11-10 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action.
Biochemistry, 40, 2001
|
|
1G7G
 
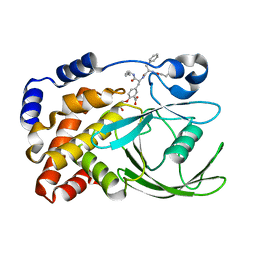 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXES WITH PNU179326 | | Descriptor: | 2-(CARBOXYMETHOXY)-5-[(2S)-2-({(2S)-2-[(3-CARBOXYPROPANOYL)AMINO] -3-PHENYLPROPANOYL}AMINO)-3-OXO-3-(PENTYLAMINO)PROPYL]BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Bleasdale, J.E, Ogg, D, Larsen, S.D. | | Deposit date: | 2000-11-10 | | Release date: | 2001-06-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action.
Biochemistry, 40, 2001
|
|
1A7S
 
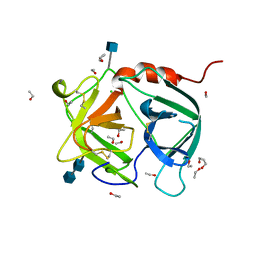 | | ATOMIC RESOLUTION STRUCTURE OF HBP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Karlsen, S, Iversen, L.F, Larsen, I.K, Flodgaard, H.J, Kastrup, J.S. | | Deposit date: | 1998-03-17 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic resolution structure of human HBP/CAP37/azurocidin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AWC
 
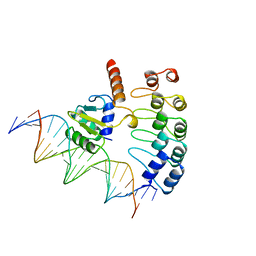 | | MOUSE GABP ALPHA/BETA DOMAIN BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*(BRU)P*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*AP*(CBR)P*AP*CP*(CBR)P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*GP*(BRU)P*GP*(BRU)P*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*AP*T)-3'), PROTEIN (GA BINDING PROTEIN ALPHA), ... | | Authors: | Batchelor, A.H, Wolberger, C. | | Deposit date: | 1997-10-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of GABPalpha/beta: an ETS domain- ankyrin repeat heterodimer bound to DNA.
Science, 279, 1998
|
|
6I9Q
 
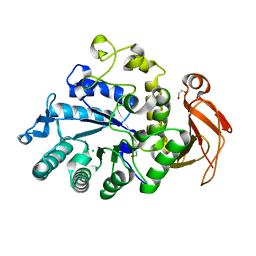 | | Structure of the mouse CD98 heavy chain ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, CHLORIDE ION | | Authors: | Schiefner, A, Deuschle, F.-C, Skerra, A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural differences between the ectodomains of murine and human CD98hc.
Proteins, 87, 2019
|
|
6KCW
 
 | | Structure of alginate lyase Aly36B | | Descriptor: | Alginate lyase, CALCIUM ION, PHOSPHATE ION | | Authors: | Dong, F, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-06-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
6KLA
 
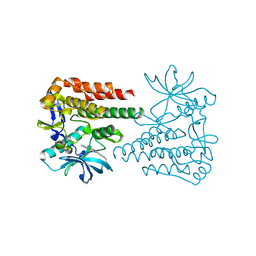 | | Crystal structure of human c-KIT kinase domain in complex with compound 15a | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-[6-(4-ethylpiperazin-1-yl)-2-methyl-pyrimidin-4-yl]-5-pyridin-4-yl-1,3-thiazol-2-amine | | Authors: | Wu, T.S, Peng, Y.H, Hsueh, C.C, Wu, S.Y. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Identification of a Multitargeted Tyrosine Kinase Inhibitor for the Treatment of Gastrointestinal Stromal Tumors and Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
2ESE
 
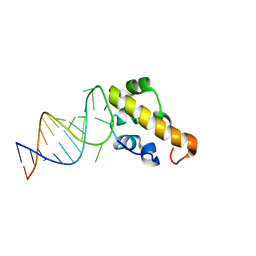 | | Structure of the SAM domain of Vts1p in complex with RNA | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*GP*CP*UP*CP*UP*GP*GP*CP*AP*GP*CP*UP*UP*UP*UP*CP*C)-3', Vts1p | | Authors: | Allain, F.H.T. | | Deposit date: | 2005-10-26 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Shape-specific recognition in the structure of the Vts1p SAM domain with RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2EVZ
 
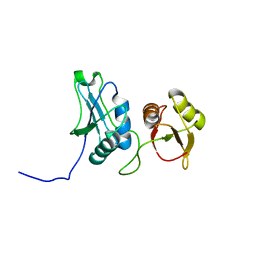 | |
2ES5
 
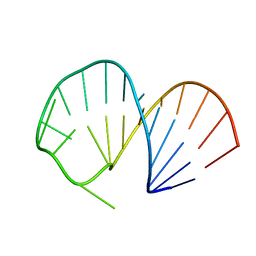 | | Structure of the SRE RNA | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*GP*CP*UP*CP*UP*GP*GP*CP*AP*GP*CP*UP*UP*UP*UP*CP*C)-3' | | Authors: | Allain, F.H.T. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Shape-specific recognition in the structure of the Vts1p SAM domain with RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
6KCV
 
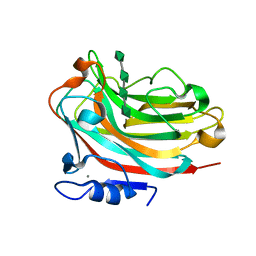 | | Structure of alginate lyase Aly36B mutant K143A/Y185A in complex with alginate tetrasaccharide | | Descriptor: | Alginate lyase, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Dong, F, Zhang, Y.Z, Chen, X.L. | | Deposit date: | 2019-06-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
2ES6
 
 | | Structure of the SAM domain of Vts1p | | Descriptor: | Vts1p | | Authors: | Allain, F.H.T. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Shape-specific recognition in the structure of the Vts1p SAM domain with RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1YPI
 
 | |
6MOY
 
 | |
