5B6K
 
 | |
6LOZ
 
 | | crystal structure of alpha-momorcharin in complex with adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LP0
 
 | | crystal structure of alpha-momorcharin in complex with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.519 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6M12
 
 | | Crystal Structure of Rnase L in complex with SU11652 | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6LOR
 
 | | crystal structure of alpha-momorcharin in complex with ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOW
 
 | | crystal structure of alpha-momorcharin in complex with GMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
7DG4
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 6 | | Descriptor: | 2,7-dimethoxy-9-(piperidin-4-ylmethylsulfanyl)acridine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DH9
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 7 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-(piperidin-4-ylmethylsulfanyl)acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHC
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 10 | | Descriptor: | 4-[bis(fluoranyl)methyl]-2,7-dimethoxy-9-(piperidin-4-ylmethylsulfanyl)acridine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHH
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 19 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [9-(azetidin-3-ylmethylsulfanyl)-2,7-dimethoxy-acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-14 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHN
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 20 | | Descriptor: | 7-methoxy-2-methylsulfanyl-9-(piperidin-4-ylmethylsulfanyl)-[1,3]thiazolo[5,4-b]quinoline, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHK
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 13 | | Descriptor: | 2-methoxy-7-phenylmethoxy-9-(piperidin-4-ylmethylsulfanyl)acridine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHV
 
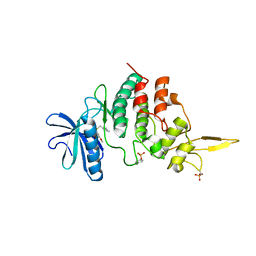 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 8 | | Descriptor: | 2,7-dimethoxy-9-(piperidin-4-ylmethylsulfanyl)acridine-4-carboxylic acid, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-17 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DHO
 
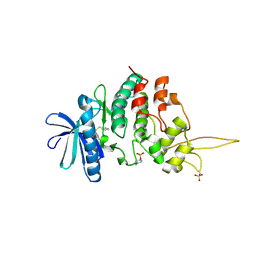 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 14 | | Descriptor: | 2-methoxy-9-(piperidin-4-ylmethylsulfanyl)-7-propan-2-yloxy-acridine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-16 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
6M13
 
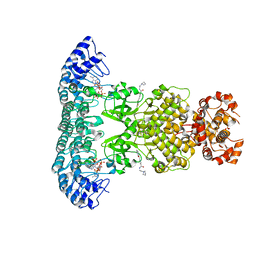 | | Crystal structure of Rnase L in complex with Toceranib | | Descriptor: | 5-[(Z)-(5-fluoranyl-2-oxidanylidene-1H-indol-3-ylidene)methyl]-2,4-dimethyl-N-(2-pyrrolidin-1-ylethyl)-1H-pyrrole-3-carboxamide, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
7DJO
 
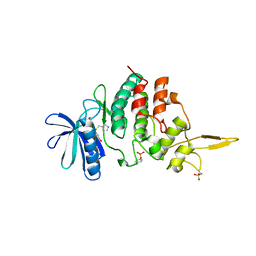 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 17 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-[[(3S)-pyrrolidin-3-yl]methylsulfanyl]acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
7DL6
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor 18 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [2,7-dimethoxy-9-[[(3R)-pyrrolidin-3-yl]methylsulfanyl]acridin-4-yl]methanol | | Authors: | Wei, T, Xiao, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Elife, 11, 2022
|
|
6LOQ
 
 | | crystal structure of alpha-momorcharin in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOY
 
 | | crystal structure of alpha-momorcharin in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
7EKQ
 
 | | CrClpP-S2c | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKO
 
 | | CrClpP-S1 | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7E7X
 
 | |
7E7Y
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E88
 
 | |
