7VUJ
 
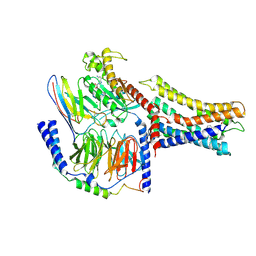 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUI
 
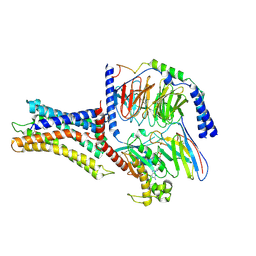 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUG
 
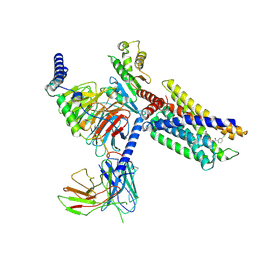 | | Cryo-EM structure of a class A orphan GPCR in complex with Gi | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
6OT1
 
 | |
6OSY
 
 | |
5TRM
 
 | |
5TRL
 
 | |
6UCE
 
 | |
6UCF
 
 | |
6D9Q
 
 | | The sulfate-bound crystal structure of HPRT (hypoxanthine phosphoribosyltransferase) | | Descriptor: | GLYCEROL, Hypoxanthine phosphoribosyltransferase, SULFATE ION | | Authors: | Satyshur, K.A, Dubiel, K, Anderson, B, Wolak, C, Keck, J.L. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Evolution of (p)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction.
Elife, 8, 2019
|
|
5E00
 
 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, GLY-VAL-TRP-ILE-ARG-THR-PRO-PRO-ALA, HLA class I histocompatibility antigen, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2015-09-26 | | Release date: | 2017-01-18 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
6D9R
 
 | | The substrate-bound crystal structure of HPRT (hypoxanthine phosphoribosyltransferase) | | Descriptor: | 1,2-ETHANEDIOL, 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAGUANINE, ... | | Authors: | Satyshur, K.A, Wolak, C, Anderson, B, Dubiel, K, Keck, J.L. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Evolution of (p)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction.
Elife, 8, 2019
|
|
6D9S
 
 | | The (p)ppGpp-bound crystal structure of HPRT (hypoxanthine phosphoribosyltransferase) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5',3'-TETRAPHOSPHATE, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Satyshur, K.A, Dubiel, K, Anderson, B, Wolak, C, Keck, J.L. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Evolution of (p)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction.
Elife, 8, 2019
|
|
8YCM
 
 | | Monomeric Human STK19 | | Descriptor: | Isoform 4 of Inactive serine/threonine-protein kinase 19, SULFATE ION | | Authors: | Li, J, Ma, X, Dong, Z. | | Deposit date: | 2024-02-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Mutations found in cancer patients compromise DNA binding of the winged helix protein STK19.
Sci Rep, 14, 2024
|
|
8WLR
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein receptor-binding domain in complex with hippopotamus ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Han, P, Yang, R.R, Li, S.H. | | Deposit date: | 2023-09-30 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Molecular basis of hippopotamus ACE2 binding to SARS-CoV-2.
J.Virol., 98, 2024
|
|
8WLO
 
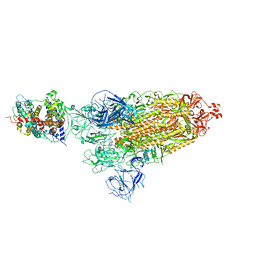 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with hippopotamus ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Yang, R.R, Li, S.H. | | Deposit date: | 2023-09-30 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Molecular basis of hippopotamus ACE2 binding to SARS-CoV-2.
J.Virol., 98, 2024
|
|
8XZB
 
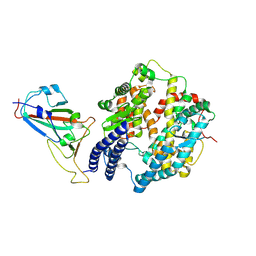 | | The structure of fox ACE2 and SARS-CoV RBD complex | | Descriptor: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | Authors: | sun, J.Q. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 2024
|
|
8W3A
 
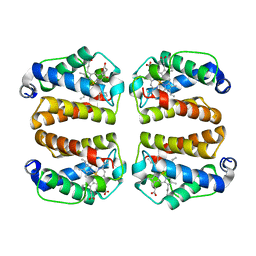 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S variant with trans heme D | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, {3-[(2R,5'R)-9',14'-diethenyl-5'-hydroxy-5',10',15',19'-tetramethyl-5-oxo-4,5-dihydro-3H-spiro[furan-2,4'-[21,22,23,24]tetraazapentacyclo[16.2.1.13,6.18,11.113,16]tetracosa[1,3(24),6,8,10,12,14,16(22),17,19]decaen]-20'-yl-kappa~4~N~21'~,N~22'~,N~23'~,N~24'~]propanoato}iron | | Authors: | Lecomte, J.T.J, Schlessman, J.L, Schultz, T.D, Siegler, M.A. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heme d formation in a Shewanella benthica hemoglobin.
J.Inorg.Biochem., 259, 2024
|
|
5H35
 
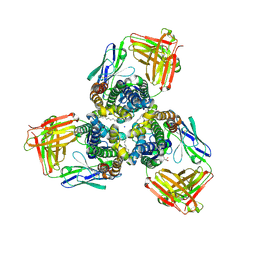 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Sulfolobus solfataricus | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
8XZD
 
 | | The structure of fox ACE2 and Omicron BF.7 RBD complex | | Descriptor: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | Authors: | sun, J.Q. | | Deposit date: | 2024-01-21 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 2024
|
|
8VSH
 
 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S variant with trans heme D | | Descriptor: | Group 1 truncated hemoglobin, {3-[(2R,5'R)-9',14'-diethenyl-5'-hydroxy-5',10',15',19'-tetramethyl-5-oxo-4,5-dihydro-3H-spiro[furan-2,4'-[21,22,23,24]tetraazapentacyclo[16.2.1.13,6.18,11.113,16]tetracosa[1,3(24),6,8,10,12,14,16(22),17,19]decaen]-20'-yl-kappa~4~N~21'~,N~22'~,N~23'~,N~24'~]propanoato}iron | | Authors: | Lecomte, J.T.J, Martinez, J.E, Schlessman, J.L, Schultz, T.D, Siegler, M.A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Heme d formation in a Shewanella benthica hemoglobin.
J.Inorg.Biochem., 259, 2024
|
|
8GVK
 
 | |
6WLG
 
 | | Ints3 C-terminal Domain | | Descriptor: | Integrator complex subunit 3 | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.111 Å) | | Cite: | Structural basis for multifunctional roles of human Ints3 C-terminal domain.
J.Biol.Chem., 296, 2020
|
|
6WPE
 
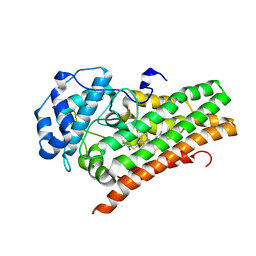 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-chloro-N-{[1-(3-chlorobenzene-1-carbonyl)-1,2,3,4-tetrahydroquinolin-6-yl]methyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Carbamate and N -Pyrimidine Mitigate Amide Hydrolysis: Structure-Based Drug Design of Tetrahydroquinoline IDO1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
5I8H
 
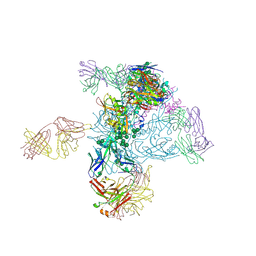 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer in Complex with V3 Loop-targeting Antibody PGT122 Fab and Fusion Peptide-targeting Antibody VRC34.01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP.664 gp120, ... | | Authors: | Xu, K, Zhou, T, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.301 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
