2GD7
 
 | |
8CZ9
 
 | |
7T6X
 
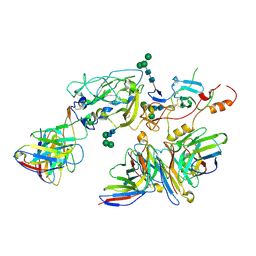 | | Cryo-EM structure of full-length hepatitis C virus E1E2 glycoprotein in complex with AR4A, AT12009, and IGH505 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AR4A Fab, ... | | Authors: | Torrents de la Pena, A, Ward, A.B, Eshun-Wilson, L, Lander, G.C. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the hepatitis C virus E1E2 glycoprotein complex.
Science, 378, 2022
|
|
5FBI
 
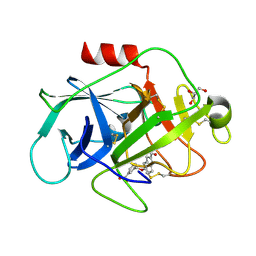 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 3b | | Descriptor: | 3-[(2-aminocarbonyl-1~{H}-indol-5-yl)oxymethyl]benzoic acid, Complement factor D, GLYCEROL | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
4UNR
 
 | | Mtb TMK in complex with compound 23 | | Descriptor: | 4-[3-cyano-2-oxo-7-(1H-pyrazol-4-yl)-5,6-dihydro-1H-benzo[h]quinolin-4-yl]benzoic acid, MAGNESIUM ION, Thymidylate kinase | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4UNN
 
 | | Mtb TMK in complex with compound 8 | | Descriptor: | 4-[3-cyano-6-(3-methoxyphenyl)-2-oxo-1H-pyridin-4-yl]benzoic acid, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-29 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
5FAH
 
 | | KALLIKREIN-7 IN COMPLEX WITH COMPOUND1 | | Descriptor: | (2~{S})-~{N}2-[2-(4-methoxyphenyl)ethyl]-~{N}1-(naphthalen-1-ylmethyl)pyrrolidine-1,2-dicarboxamide, ACETATE ION, Kallikrein-7 | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FCK
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[[(1~{R})-1-(3-chloranyl-2-fluoranyl-phenyl)ethyl]carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]pyrazolo[3,4-c]pyridine-3-carboxamide, Complement factor D, SULFATE ION | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
4UNP
 
 | | Mtb TMK in complex with compound 34 | | Descriptor: | 5-methyl-7-propyl-1,6-naphthyridin-2(1H)-one, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4UNQ
 
 | | Mtb TMK in complex with compound 36 | | Descriptor: | 4-[(R)-methylsulfinyl]-2-oxo-6-[3-(trifluoromethoxy)phenyl]-1,2-dihydropyridine-3-carbonitrile, SODIUM ION, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4UNS
 
 | | Mtb TMK in complex with compound 40 | | Descriptor: | N-[4-(3-CYANO-7-ETHYL-5-METHYL-2-OXO-1H-1,6-NAPHTHYRIDIN-4-YL)PHENYL]METHANESULFONAMIDE, SODIUM ION, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
5FBE
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND2 | | Descriptor: | Complement factor D, GLYCEROL, methyl 2-[[[(2~{S})-2-[[3-(trifluoromethyloxy)phenyl]carbamoyl]pyrrolidin-1-yl]carbonylamino]methyl]benzoate | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FCR
 
 | | MOUSE COMPLEMENT FACTOR D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Complement factor D, DIMETHYL SULFOXIDE, ... | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
1YX3
 
 | | NMR structure of Allochromatium vinosum DsrC: Northeast Structural Genomics Consortium target OP4 | | Descriptor: | hypothetical protein DsrC | | Authors: | Cort, J.R, Dahl, C, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation.
J.Mol.Biol., 382, 2008
|
|
7WHG
 
 | |
7WHF
 
 | | Heimdallarchaeota gelsolin (2DGel) bound to rabbit actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Robinson, R.C, Akil, C. | | Deposit date: | 2021-12-30 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical evidence for the emergence of a calcium-regulated actin cytoskeleton prior to eukaryogenesis
Commun Biol, 5, 2022
|
|
7O6U
 
 | |
7O6T
 
 | | Crystal structure of the polymerising VEL domain of VIN3 (R556D I575D mutant) | | Descriptor: | MAGNESIUM ION, Protein VERNALIZATION INSENSITIVE 3 | | Authors: | Fiedler, M, Franco-Echevarria, E, Dean, C, Bienz, M. | | Deposit date: | 2021-04-12 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Head-to-tail polymerization by VEL proteins underpins cold-induced Polycomb silencing in flowering control.
Cell Rep, 41, 2022
|
|
7O6W
 
 | | Crystal structure of (the) VEL1 VEL polymerising domain (I664D mutant) | | Descriptor: | PHOSPHATE ION, VIN3-like protein 2 | | Authors: | Fiedler, M, Franco-Echevarria, E, Dean, C, Bienz, M. | | Deposit date: | 2021-04-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Head-to-tail polymerization by VEL proteins underpins cold-induced Polycomb silencing in flowering control.
Cell Rep, 41, 2022
|
|
7O6V
 
 | |
7OQV
 
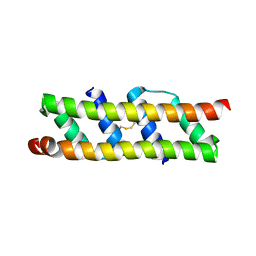 | |
7AK0
 
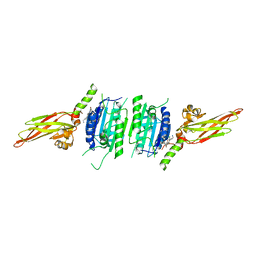 | | Human MALT1(329-729) in complex with a chromane urea containing inhibitor | | Descriptor: | 1-[4-[4-(aminomethyl)pyrazol-1-yl]-3-chloranyl-phenyl]-3-[(3~{R})-6-bromanyl-3,4-dihydro-2~{H}-chromen-3-yl]urea, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Discovery of Potent, Highly Selective, and In Vivo Efficacious, Allosteric MALT1 Inhibitors by Iterative Scaffold Morphing.
J.Med.Chem., 63, 2020
|
|
8QPD
 
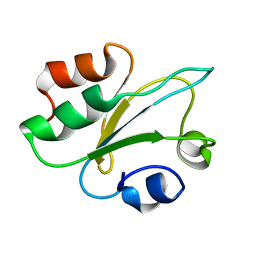 | | Structure of thioredoxin m from pea | | Descriptor: | Thioredoxin M-type, chloroplastic | | Authors: | Neira, J.L, Camara Artigas, A. | | Deposit date: | 2023-10-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure, dynamics and binding of thioredoxin m from Pisum sativum.
Int.J.Biol.Macromol., 262, 2024
|
|
5MDR
 
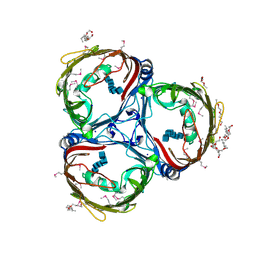 | | Crystal structure of in vitro folded Chitoporin VhChip from Vibrio harveyi in complex with chitohexaose | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitoporin, ... | | Authors: | Zahn, M, van den Berg, B. | | Deposit date: | 2016-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for chitin acquisition by marine Vibrio species.
Nat Commun, 9, 2018
|
|
5MDO
 
 | |
