8BHS
 
 | | GABA-A receptor a5 homomer - a5V3 - RO4938581 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[bis(fluoranyl)methyl]-15-bromanyl-2,4,8,9,11-pentazatetracyclo[11.4.0.0^{2,6}.0^{8,12}]heptadeca-1(13),3,5,9,11,14,16-heptaene, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BGI
 
 | | GABA-A receptor a5 homomer - a5V1 - Flumazenil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, Pregnanolone, ... | | Authors: | Miller, P.S, Malinauskas, T.M, Omari, K.E, Aricescu, A.R. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHA
 
 | | GABA-A receptor a5 homomer - a5V3 - Basmisanil - HR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Basmisanil, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Malinauskas, T.M, Wahid, A.A, Hardwick, S.W, Chirgadze, D.Y, Miller, P.S. | | Deposit date: | 2022-10-30 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHR
 
 | | GABA-A receptor a5 homomer - a5V3 - RO7015738 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[[5-methyl-3-(6-methylpyridin-3-yl)-1,2-oxazol-4-yl]methoxy]-~{N}-[(2~{S})-1-oxidanylpentan-2-yl]pyridine-3-carboxamide, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Kasaragod, V.B. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
6FKK
 
 | | Drosophila Semaphorin 1b, extracellular domains 1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MIP07328p, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rozbesky, D, Harlos, K, Jones, E.Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
5I1H
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV3-20 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1E
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV1-39 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I18
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV1-69/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I16
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV1-69/IGKV3-11 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1K
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV5-51/IGKV3-20 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1C
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-23/IGKV3-20 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FAB HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1G
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV3-11 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I19
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-23/IGKV1-39 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1J
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV5-51/IGKV3-11 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I17
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV1-69/IGKV3-20 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
7JGT
 
 | | Crystal Structure of FN3tt | | Descriptor: | Fibronectin type-III domain-containing protein | | Authors: | Luo, J, Malia, T.J. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
5I1D
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-23/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1A
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-23/IGKV3-11 | | Descriptor: | ACETATE ION, CNTO388 HEAVY CHAIN, CNTO388 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1L
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV5-51/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I15
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV1-69/IGKV1-39 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FAB HEAVY CHAIN, FAB LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5I1I
 
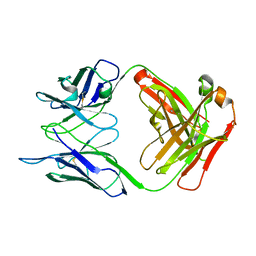 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
6XS6
 
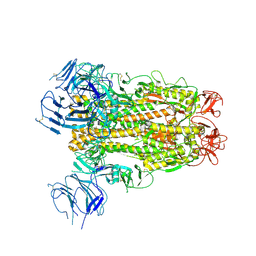 | | SARS-CoV-2 Spike D614G variant, minus RBD | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Egri, S.B, Dudkina, N, Luban, J, Shen, K. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant.
Cell, 183, 2020
|
|
3I84
 
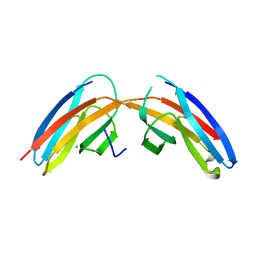 | |
