7F1S
 
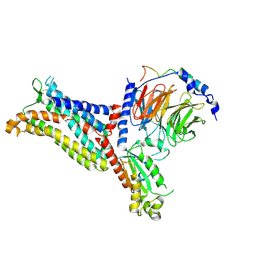 | | Cryo-EM structure of the apo chemokine receptor CCR5 in complex with Gi | | Descriptor: | C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F58
 
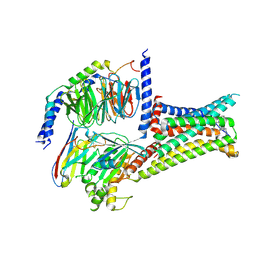 | | Cryo-EM structure of THIQ-MC4R-Gs_Nb35 complex | | Descriptor: | (3R)-N-[(2R)-3-(4-chlorophenyl)-1-[4-cyclohexyl-4-(1,2,4-triazol-1-ylmethyl)piperidin-1-yl]-1-oxidanylidene-propan-2-yl]-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F55
 
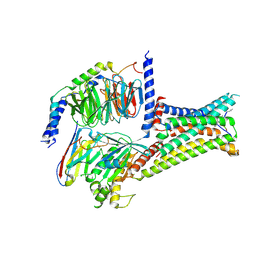 | | Cryo-EM structure of bremelanotide-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F54
 
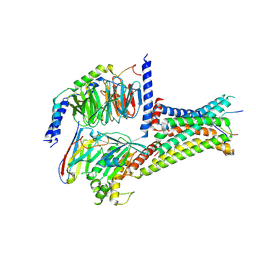 | | Cryo-EM structure of afamelanotide-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F53
 
 | | Cryo-EM structure of a-MSH-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7CQ1
 
 | |
7XB6
 
 | |
7EDK
 
 | |
3CHT
 
 | |
5XP6
 
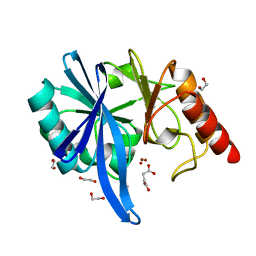 | | native structure of NDM-1 crystallized at pH5.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G, Lai, J, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
5XP9
 
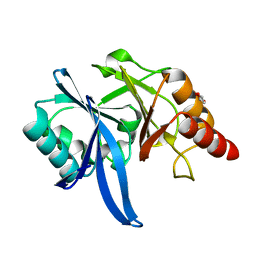 | | Crystal structure of Bismuth bound NDM-1 | | Descriptor: | Bismuth(III) ION, GLYCEROL, Metallo-beta-lactamase type 2 | | Authors: | Zhang, H, Ma, G, Lai, T.P, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
3S6T
 
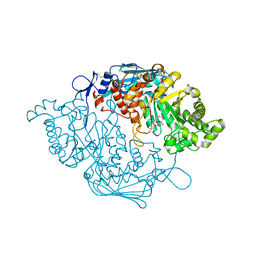 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 V327G complexed with PUGNAc | | Descriptor: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Zhang, H, Liu, T, Yang, Q, Shen, X. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases.
Biochem.J., 438, 2011
|
|
5XAG
 
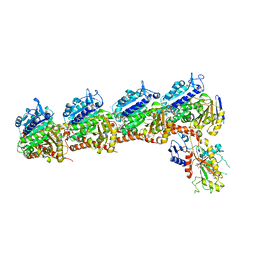 | | Crystal structure of tubulin-stathmin-TTL-Compound Z2 complex | | Descriptor: | (3~{R},4~{R})-3-(hydroxymethyl)-4-(4-methoxy-3-oxidanyl-phenyl)-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
6LJ1
 
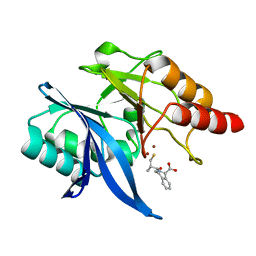 | |
6LJ2
 
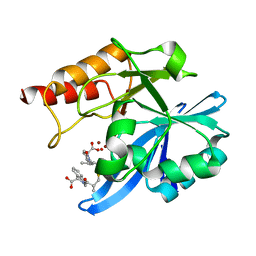 | | Crystal structure of NDM-1 in complex with heterodimer of D-captopril derivative wss02127 stereoisomer | | Descriptor: | (1R)-2-[(2S)-2-methyl-3-sulfanyl-propanoyl]-3,4-dihydro-1H-isoquinoline-1-carboxylic acid, (1S)-2-[(2S)-2-methyl-3-sulfanyl-propanoyl]-3,4-dihydro-1H-isoquinoline-1-carboxylic acid, HYDROXIDE ION, ... | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJ5
 
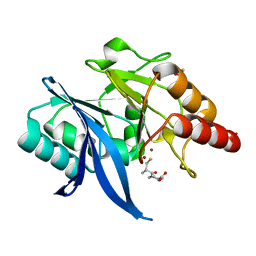 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss04145 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJ0
 
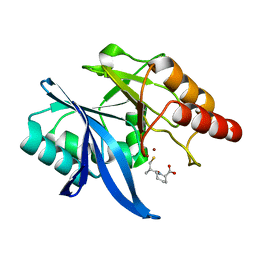 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss02122 | | Descriptor: | (2R)-1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidine-2-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJ6
 
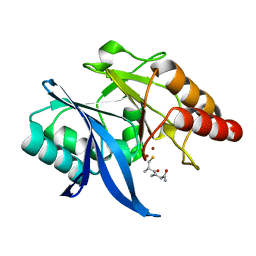 | |
6LJ4
 
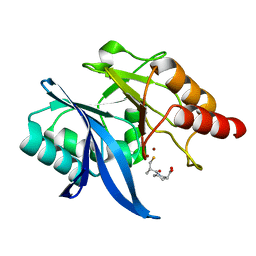 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss04146 | | Descriptor: | (3S)-1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidine-3-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJ8
 
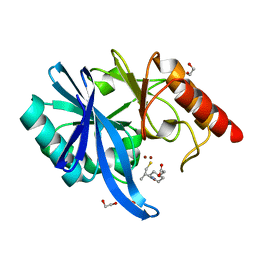 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss04134 | | Descriptor: | 1,2-ETHANEDIOL, 2-[1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidin-4-yl]ethanoic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LIP
 
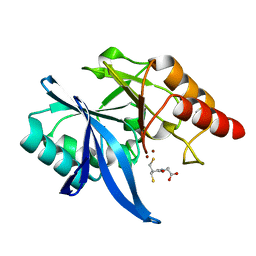 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss0218 | | Descriptor: | (2R)-1-[3-sulfanyl-2-(sulfanylmethyl)propanoyl]pyrrolidine-2-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LIZ
 
 | |
6LJ7
 
 | |
6LL9
 
 | |
5XOM
 
 | | Hydra Fam20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosaminoglycan xylosylkinase | | Authors: | Xiao, J, Zhang, H. | | Deposit date: | 2017-05-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
