1ZKP
 
 | | 1.5A Resolution Crystal Structure of a Metallo Beta Lactamase Family Protein, the ELAC Homolgue of Bacillus anthracis, a Putative Ribonuclease | | Descriptor: | CHLORIDE ION, SODIUM ION, ZINC ION, ... | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | 1.5A Resolution Crystal Structure of a Metallo Beta Lactamase Family
Protein, the ELAC Homolgue of Bacillus anthracis, a Putative Ribonuclease
To be Published
|
|
2ATR
 
 | | Acetyltransferase, GNAT family protein SP0256 from Streptococcus pneumoniae TIGR4 | | Descriptor: | acetyltransferase, GNAT family | | Authors: | Binkowski, T.A, Hatzos, C, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-25 | | Release date: | 2005-10-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Acetyltransferase, GNAT family protein SP0256 from Streptococcus pneumoniae
To be Published
|
|
2GTS
 
 | | Structure of Protein of Unknown Function HP0062 from Helicobacter pylori | | Descriptor: | hypothetical protein HP0062 | | Authors: | Binkowski, T.A, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hypothetical protein HP0062 from Helicobacter pylori
To be Published
|
|
1YLE
 
 | |
1YX2
 
 | |
1YLM
 
 | |
1YOY
 
 | |
1YLL
 
 | |
1YLX
 
 | |
1YWQ
 
 | |
1YZH
 
 | |
4TX9
 
 | | Crystal structure of HisAp from Streptomyces sviceus with degraded ProFAR | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Phosphoribosyl isomerase A, SULFATE ION | | Authors: | Michalska, K, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4DGH
 
 | | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution | | Descriptor: | GLYCEROL, IODIDE ION, POTASSIUM ION, ... | | Authors: | Keller, J.P, Chang, C, Marshall, N, Bearden, J, Dallos, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution
To be Published
|
|
4W66
 
 | |
5CRF
 
 | | Structure of the penicillin-binding protein PonA1 from Mycobacterium Tuberculosis | | Descriptor: | PHOSPHATE ION, Penicillin-binding protein 1A | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-22 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
4LMI
 
 | |
4LLE
 
 | |
4LLC
 
 | | The crystal structure of R60E mutant of the histidine kinase (KinB) sensor domain from Pseudomonas aeruginosa PA01 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Probable two-component sensor, ... | | Authors: | Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of R60E mutant of the histidine kinase (KinB) sensor domain from Pseudomonas aeruginosa PA01
To be Published
|
|
5CXW
 
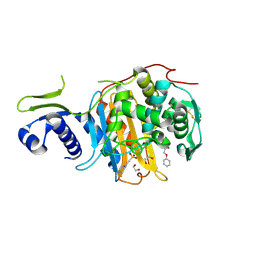 | | Structure of the PonA1 protein from Mycobacterium Tuberculosis in complex with penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
4LJS
 
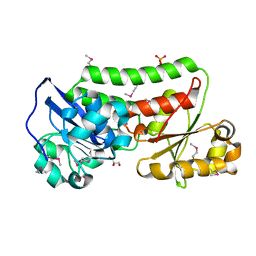 | | The crystal structure of a periplasmic binding protein from Veillonella parvula DSM 2008 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Periplasmic binding protein | | Authors: | Tan, K, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-05 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | The crystal structure of a periplasmic binding protein from Veillonella parvula DSM 2008
To be Published
|
|
4W97
 
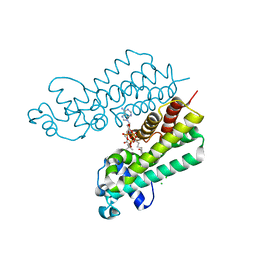 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
5CR9
 
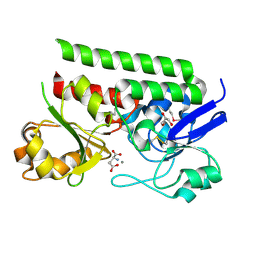 | | Crystal structure of ABC-type Fe3+-hydroxamate transport system from Saccharomonospora viridis DSM 43017 | | Descriptor: | ABC-type Fe3+-hydroxamate transport system, periplasmic component, GLUTAMIC ACID, ... | | Authors: | Nocek, B, Cuff, M, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ABC-type Fe3+-hydroxamate transport system from Saccharomonospora viridis DSM 43017
To Be Published
|
|
5DDG
 
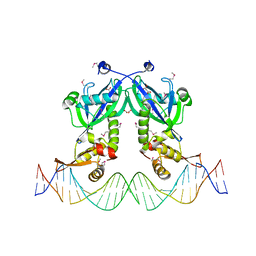 | | The structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with target double strand DNA | | Descriptor: | DNA (27-MER), FORMIC ACID, MALONIC ACID, ... | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5DD4
 
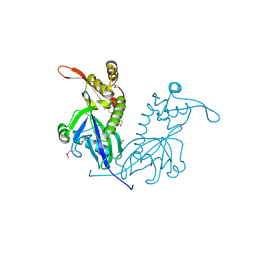 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, TRANSCRIPTIONAL REGULATOR AraR | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
4W9R
 
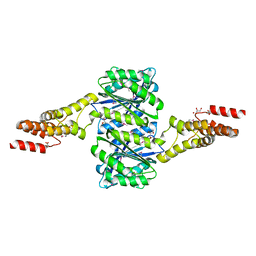 | | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271
To Be Published
|
|
