7KBR
 
 | | Co-crystal structure of alpha glucosidase with compound 10 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7L9E
 
 | | Crystal structure of apo-alpha glucosidase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-01-03 | | Release date: | 2021-12-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KRY
 
 | | Co-crystal structure of alpha glucosidase with compound 11 | | Descriptor: | (1S,2S,3R,4S,5S)-5-({6-[(4-azido-2-nitrophenyl)amino]hexyl}amino)-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
3S5L
 
 | | Crystal structure of CD4 mutant bound to HLA-DR1 | | Descriptor: | GLYCEROL, HA peptide, HLA class II histocompatibility antigen DR beta chain, ... | | Authors: | Li, Y. | | Deposit date: | 2011-05-23 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S4S
 
 | | Crystal structure of CD4 mutant bound to HLA-DR1 | | Descriptor: | GLYCEROL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Li, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4PO4
 
 | |
5EUO
 
 | | PF6-M1-HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Yang, X. | | Deposit date: | 2015-11-18 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PF6-M1-HLA-A2 complex
To Be Published
|
|
2NTS
 
 | | Crystal Structure of SEK-hVb5.1 | | Descriptor: | Staphylococcal enterotoxin K, TRBC1 protein | | Authors: | Gunther, S, Varma, A.K, Moza, B, Sundberg, E.J. | | Deposit date: | 2006-11-08 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel loop domain in superantigens extends their T cell receptor recognition site
J.Mol.Biol., 371, 2007
|
|
2NTT
 
 | | Crystal Structure of SEK | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Staphylococcal enterotoxin K | | Authors: | Gunther, S, Varma, A.K, Moza, B, Sundberg, E.J. | | Deposit date: | 2006-11-08 | | Release date: | 2007-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | A novel loop domain in superantigens extends their T cell receptor recognition site
J.Mol.Biol., 371, 2007
|
|
8DNT
 
 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
9G3Q
 
 | | Chitinase-like protein AgBR1 from Aedes aegypti | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAEL001965-PA, D-MALATE | | Authors: | Abrescia, N.G.A, Martinez-Castillo, A. | | Deposit date: | 2024-07-12 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and functional significance of Aedes aegypti AgBR1 flavivirus immunomodulator.
J.Virol., 99, 2025
|
|
1JA3
 
 | | Crystal Structure of the Murine NK Cell Inhibitory Receptor Ly-49I | | Descriptor: | MHC class I recognition receptor Ly49I | | Authors: | Dimasi, N, Sawicki, W.M, Reineck, L.A, Li, Y, Natarajan, K, Murgulies, D.H, Mariuzza, A.R. | | Deposit date: | 2001-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Ly49I natural killer cell receptor reveals variability in dimerization mode within the Ly49 family.
J.Mol.Biol., 320, 2002
|
|
1HQR
 
 | | CRYSTAL STRUCTURE OF A SUPERANTIGEN BOUND TO THE HIGH-AFFINITY, ZINC-DEPENDENT SITE ON MHC CLASS II | | Descriptor: | HLA-DR ALPHA CHAIN, HLA-DR BETA CHAIN, MYELIN BASIC PROTEIN, ... | | Authors: | Li, Y, Li, H, Dimasi, N, Schlievert, P, Mariuzza, R. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a superantigen bound to the high-affinity, zinc-dependent site on MHC class II.
Immunity, 14, 2001
|
|
3BYY
 
 | |
3BZD
 
 | |
3BVG
 
 | |
3BVM
 
 | |
3BVZ
 
 | |
3BYT
 
 | |
1FV1
 
 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
8FSJ
 
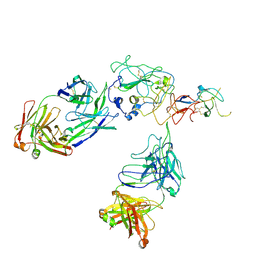 | | Cryo-EM structure of engineered hepatitis C virus E1E2 ectodomain in complex with antibodies AR4A, HEPC74, and IGH520 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AR4A heavy chain, ... | | Authors: | Metcalf, M.C, Ofek, G. | | Deposit date: | 2023-01-10 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.
Nat Commun, 14, 2023
|
|
4IOP
 
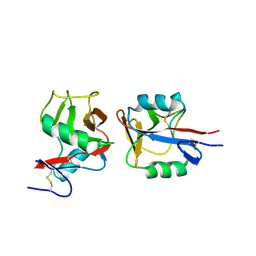 | | Crystal structure of NKp65 bound to its ligand KACL | | Descriptor: | C-type lectin domain family 2 member A, Killer cell lectin-like receptor subfamily F member 2, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y. | | Deposit date: | 2013-01-08 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of NKp65 bound to its keratinocyte ligand reveals basis for genetically linked recognition in natural killer gene complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IS6
 
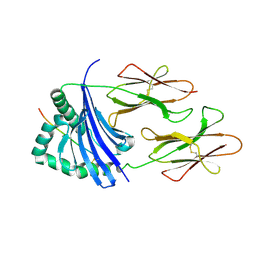 | | Crystal structure of HLA-DR4 bound to GP100 peptide | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-4 beta chain, ... | | Authors: | Li, Y. | | Deposit date: | 2013-01-16 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Altered MHC Class II-Restricted Peptide Ligands with Heterogeneous Immunogenicity.
J.Immunol., 191, 2013
|
|
4Z8I
 
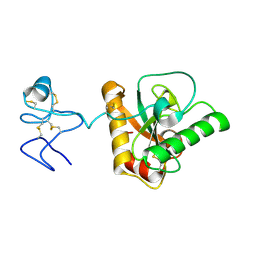 | | Crystal structure of Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | ZINC ION, peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Luo, M, Cao, D.D, Chi, C.B, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
1A7N
 
 | |
