5UK0
 
 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
2KXY
 
 | | Solution structure of SuR18C from Streptococcus thermophilus. Northeast Structural Genomics Consortium Target SuR18C | | Descriptor: | Uncharacterized protein | | Authors: | Liu, Y, Lee, H, Belote, R, Ciccosanti, C, Hamilton, K, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SuR18C from Streptococcus thermophilus. Northeast Structural Genomics Consortium Target SuR18C
To be Published
|
|
5UZA
 
 | | Adenine riboswitch aptamer domain labelled with iodo-uridine by position-selective labelling of RNA (PLOR) | | Descriptor: | ADENINE, MAGNESIUM ION, RNA (71-MER) | | Authors: | Liu, Y, Stagno, J.R, Wang, Y.-X. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA.
Nat Protoc, 13, 2018
|
|
2L8O
 
 | | Solution structure of Chr148 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target Chr148 | | Descriptor: | Activator of Hsp90 ATPase homologue 1-like C-terminal domain-containing protein | | Authors: | Liu, Y, Lee, D, Ciccosanti, C, Nair, L, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Chr148 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target Chr148
To be Published
|
|
5UK1
 
 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
2KSQ
 
 | | The myristoylated yeast ARF1 in a GTP and bicelle bound conformation | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Liu, Y, Kahn, R, Prestegard, J. | | Deposit date: | 2010-01-12 | | Release date: | 2010-07-07 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Dynamic structure of membrane-anchored Arf*GTP.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3SSJ
 
 | |
3VAC
 
 | |
3THQ
 
 | |
3SW6
 
 | |
3TEJ
 
 | |
8WHA
 
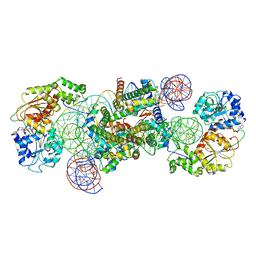 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH5
 
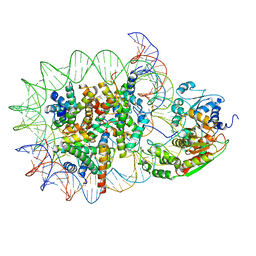 | | Structure of DDM1-nucleosome complex in the apo state | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHB
 
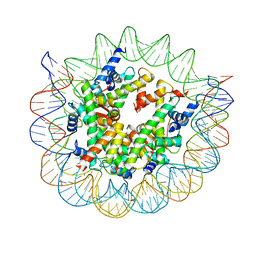 | | Structure of nucleosome core particle of Arabidopsis thaliana | | Descriptor: | DNA (antisense strand), DNA (sense strand), Histone H2A.6, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-23 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH8
 
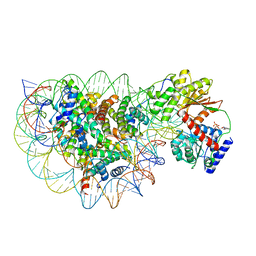 | | Structure of DDM1-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, DNA (antisense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH9
 
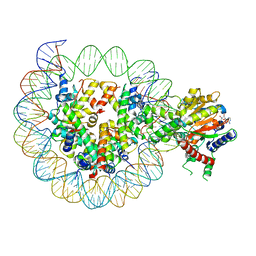 | | Structure of DDM1-nucleosome complex in ADP-BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
7VKH
 
 | |
7VKG
 
 | |
5YVX
 
 | | Crystal structure of SDG8 CW domain in complex with H3K4me1 peptide | | Descriptor: | H3K4me1, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Liu, Y, Huang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Uncovering the mechanistic basis for specific recognition of monomethylated H3K4 by the CW domain ofArabidopsishistone methyltransferase SDG8.
J. Biol. Chem., 293, 2018
|
|
6JPI
 
 | |
8H43
 
 | |
7VYW
 
 | |
7VZ2
 
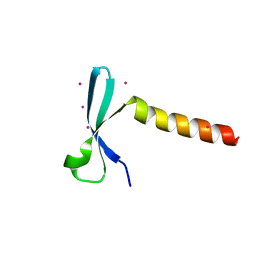 | | Crystal structure of chromodomain of Arabidopsis LHP1 | | Descriptor: | Chromo domain-containing protein LHP1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Min, J. | | Deposit date: | 2021-11-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the recognition of methylated histone H3 by the Arabidopsis LHP1 chromodomain.
J.Biol.Chem., 298, 2022
|
|
7WDL
 
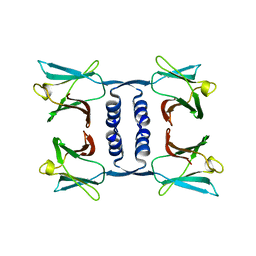 | | Fungal immunomodulatory protein FIP-nha | | Descriptor: | Fungal immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties.
Int.J.Biol.Macromol., 213, 2022
|
|
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
