4XP9
 
 | | X-ray structure of Drosophila dopamine transporter bound to psychostimulant D-amphetamine | | Descriptor: | (2S)-1-phenylpropan-2-amine, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XPA
 
 | | X-ray structure of Drosophila dopamine transporter bound to 3,4dichlorophenethylamine | | Descriptor: | 2-(3,4-dichlorophenyl)ethanamine, Antibody fragment heavy chain-protein, 9D5-heavy chain, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
3H9V
 
 | | Crystal structure of the ATP-gated P2X4 ion channel in the closed, apo state at 3.1 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GADOLINIUM ATOM, P2X purinoceptor | | Authors: | Kawate, T, Michel, J.C, Gouaux, E. | | Deposit date: | 2009-04-30 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the ATP-gated P2X(4) ion channel in the closed state.
Nature, 460, 2009
|
|
1MQH
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Bromo-Willardiine at 1.8 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MQI
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Fluoro-Willardiine at 1.35 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MY0
 
 | | crystal titration experiments (AMPA co-crystals soaked in 100 nM BrW) | | Descriptor: | GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
1MXU
 
 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) in complex with bromo-willardiine (Control for the crystal titration experiments) | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
1MXV
 
 | | crystal titration experiments (AMPA co-crystals soaked in 10 mM BrW) | | Descriptor: | GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
1MY2
 
 | | crystal titration experiment (AMPA complex control) | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the function, conformational plasticity, and dimer-dimer contacts of the GluR2 ligand-binding core: studies of 5-substituted willardiines and GluR2 S1S2 in the crystal.
Biochemistry, 42, 2003
|
|
1MXY
 
 | | crystal titration experiments (AMPA co-crystals soaked in 10 uM BrW) | | Descriptor: | GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the function, conformational plasticity, and dimer-dimer contacts of the GluR2 ligand-binding core: studies of 5-substituted willardiines and GluR2 S1S2 in the crystal.
Biochemistry, 42, 2003
|
|
1MXW
 
 | | crystal titration experiments (AMPA co-crystals soaked in 1 mM BrW) | | Descriptor: | GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the function, conformational plasticity, and dimer-dimer contacts of the GluR2 ligand-binding core: studies of 5-substituted willardiines and GluR2 S1S2 in the crystal.
Biochemistry, 42, 2003
|
|
1MXZ
 
 | | crystal titration experiments (AMPA co-crystals soaked in 1 uM BrW) | | Descriptor: | GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the function, conformational plasticity, and dimer-dimer contacts of the GluR2 ligand-binding core: studies of 5-substituted willardiines and GluR2 S1S2 in the crystal.
Biochemistry, 42, 2003
|
|
3I5D
 
 | | Crystal structure of the ATP-gated P2X4 ion channel in the closed, apo state at 3.5 Angstroms (R3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2X purinoceptor | | Authors: | Kawate, T, Michel, J.C, Gouaux, E. | | Deposit date: | 2009-07-05 | | Release date: | 2009-08-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of the ATP-gated P2X(4) ion channel in the closed state.
Nature, 460, 2009
|
|
1MY4
 
 | | crystal structure of glutamate receptor ligand-binding core in complex with iodo-willardiine in the Zn crystal form | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
5I74
 
 | | X-ray structure of the ts3 human serotonin transporter complexed with Br-citalopram at the central site | | Descriptor: | (1S)-1-(4-bromophenyl)-1-[3-(dimethylamino)propyl]-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.395 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
5I71
 
 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central site | | Descriptor: | (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
3GI9
 
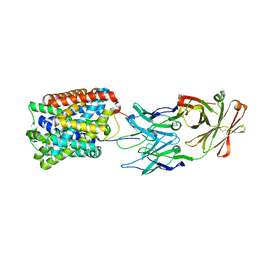 | | Crystal Structure of ApcT Transporter Bound to 7F11 Monoclonal Fab Fragment | | Descriptor: | 7F11 Anti-ApcT Monoclonal Fab Heavy Chain, 7F11 Anti-ApcT Monoclonal Fab Light Chain, Uncharacterized protein MJ0609 | | Authors: | Shaffer, P.L, Goehring, A.S, Shankaranarayanan, A, Gouaux, E, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and mechanism of a na+-independent amino Acid transporter.
Science, 325, 2009
|
|
5IPT
 
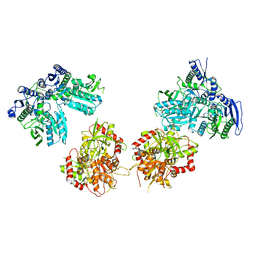 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 5 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
3GI8
 
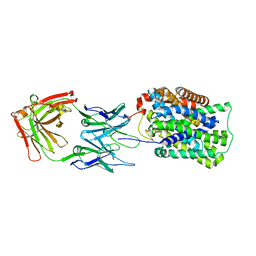 | | Crystal Structure of ApcT K158A Transporter Bound to 7F11 Monoclonal Fab Fragment | | Descriptor: | 7F11 Anti-ApcT Monoclonal Fab Heavy Chain, 7F11 Anti-ApcT Monoclonal Fab Light Chain, Uncharacterized protein MJ0609 | | Authors: | Shaffer, P.L, Goehring, A.S, Shankaranarayanan, A, Gouaux, E, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure and mechanism of a na+-independent amino Acid transporter.
Science, 325, 2009
|
|
5I73
 
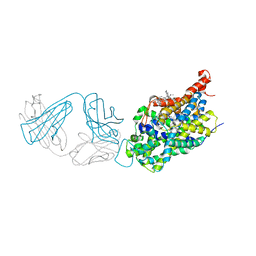 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central and allosteric sites | | Descriptor: | (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
5IOV
 
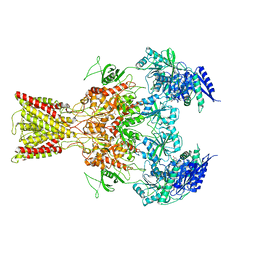 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine/Ro25-6981-bound conformation | | Descriptor: | 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPV
 
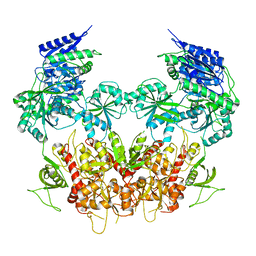 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 1 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, McHaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.25 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5I6Z
 
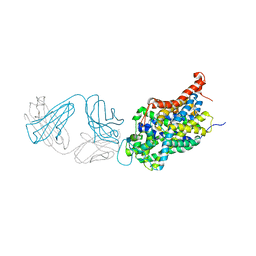 | | X-ray structure of the ts2 human serotonin transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, heavy chain, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
5IPR
 
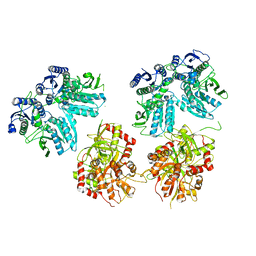 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 3 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
3GIA
 
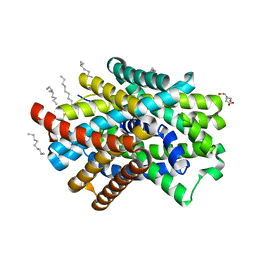 | | Crystal Structure of ApcT Transporter | | Descriptor: | BICINE, DECANE, Uncharacterized protein MJ0609 | | Authors: | Shaffer, P.L, Goehring, A.S, Shankaranarayanan, A, Gouaux, E, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and mechanism of a na+-independent amino Acid transporter.
Science, 325, 2009
|
|
