6WUX
 
 | |
5MQQ
 
 | |
7SOL
 
 | | Crystal Structures of the bispecific ubiquitin/FAT10 activating enzyme, Uba6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Ubiquitin, ... | | Authors: | Olsen, S.K, Gao, F, Lv, Z. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.25000644 Å) | | Cite: | Crystal structures reveal catalytic and regulatory mechanisms of the dual-specificity ubiquitin/FAT10 E1 enzyme Uba6.
Nat Commun, 13, 2022
|
|
8G46
 
 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2 | | Descriptor: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DET1- and DDB1-associated protein 1, ... | | Authors: | Ma, M.W, Hunkeler, M, Jin, C.Y, Fischer, E.S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Template-assisted covalent modification of DCAF16 underlies activity of BRD4 molecular glue degraders.
Biorxiv, 2023
|
|
6XN0
 
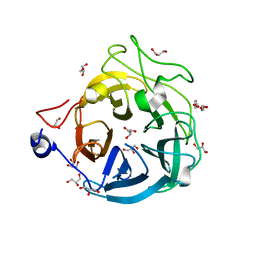 | | Crystal structure of GH43_1 enzyme from Xanthomonas citri | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
7P4U
 
 | |
5L16
 
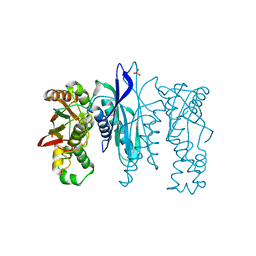 | | Crystal Structure of N-terminus truncated selenophosphate synthetase from Leishmania major | | Descriptor: | Putative selenophosphate synthetase, SULFATE ION | | Authors: | Faim, L.M, Silva, I.R, Pereira, H.M, Dias, M.B, Silva, M.T.A, Brandao-Neto, J, Thiemann, O.H. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Trypanosomatid selenophosphate synthetase structure, function and interaction with selenocysteine lyase.
Plos Negl Trop Dis, 14, 2020
|
|
2YFE
 
 | |
2YPI
 
 | |
5M6Q
 
 | | Crystal Structure of Kutzneria albida transglutaminase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Steffen, W, Benz, J, Rudolph, M.G. | | Deposit date: | 2016-10-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a microbial transglutaminase enabling highly site-specific labeling of proteins.
J. Biol. Chem., 292, 2017
|
|
5TBO
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM421 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[6-(trifluoromethyl)pyridin-3-yl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M. | | Deposit date: | 2016-09-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | A Triazolopyrimidine-Based Dihydroorotate Dehydrogenase Inhibitor with Improved Drug-like Properties for Treatment and Prevention of Malaria.
ACS Infect Dis, 2, 2016
|
|
1FY1
 
 | | [R23S,F25E]HBP, A MUTANT OF HUMAN HEPARIN BINDING PROTEIN (CAP37) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHANOL, ... | | Authors: | Kastrup, J.S, Linde, V, Pedersen, A.K, Stoffer, B, Iversen, L.F, Larsen, I.K, Rasmussen, P.B, Flodgaard, H.J, Bjorn, S.E. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two mutants of human heparin binding protein (CAP37): toward the understanding of the nature of lipid A/LPS and BPTI binding.
Proteins, 42, 2001
|
|
1FY3
 
 | | [G175Q]HBP, A mutant of human heparin binding protein (CAP37) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kastrup, J.S, Linde, V, Pedersen, A.K, Stoffer, B, Iversen, L.F, Larsen, I.K, Rasmussen, P.B, Flodgaard, H.J, Bjorn, S.E. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Two mutants of human heparin binding protein (CAP37): toward the understanding of the nature of lipid A/LPS and BPTI binding.
Proteins, 42, 2001
|
|
7PON
 
 | |
7PNO
 
 | | C terminal domain of Nipah Virus Phosphoprotein fused to the Ntail alpha more of the Nucleoprotein. | | Descriptor: | Phosphoprotein, alpha MoRE of Nipah virus Nucleoprotein tail | | Authors: | Bourhis, J.M, Yabukaski, F, Tarbouriech, N, Jamin, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-04-20 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Dynamics of the C-terminal X Domain of Nipah and Hendra Viruses Controls the Attachment to the C-terminal Tail of the Nucleocapsid Protein.
J.Mol.Biol., 434, 2022
|
|
6KZK
 
 | |
7RU7
 
 | | Crystal structure of BtrK, a decarboxylase involved in butirosin biosynthesis | | Descriptor: | DI(HYDROXYETHYL)ETHER, L-glutamyl-[BtrI acyl-carrier protein] decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Arenas, L.A.R, Paiva, F.C.R, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of BtrK, a decarboxylase involved in the (S)-4-amino-2-hydroxybutyrate (AHBA) formation during butirosin biosynthesis
J.Mol.Struct., 1267, 2022
|
|
8UUO
 
 | | Prototypic SARS-CoV-2 spike (containing V417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUM
 
 | | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUN
 
 | | Prototypic SARS-CoV-2 spike (containing V417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUL
 
 | | Prototypic SARS-CoV-2 spike (containing K417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
6XN2
 
 | | Crystal structure of the GH43_1 enzyme from Xanthomonas citri complexed with xylotriose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
6XN1
 
 | | Crystal structure of the GH43_1 enzyme from Xanthomonas citri complexed with xylose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
4YNK
 
 | | Crystal structure of vitamin D receptor ligand binding domain complexed with a 19-norvitamin D compound | | Descriptor: | (1R,3R,7E,17beta)-17-{(5S)-5-hydroxy-5-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-1-yl]penta-1,3-diyn-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Coactivator peptide drip from cDNA FLJ50196, highly similar to Peroxisome proliferator-activated receptor-binding protein, ... | | Authors: | Watarai, Y, Ikura, T, Ito, N. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, Biological Activities, and X-ray Crystal Structural Analysis of 25-Hydroxy-25(or 26)-adamantyl-17-[20(22),23-diynyl]-21-norvitamin D Compounds
J.Med.Chem., 58, 2015
|
|
4U0V
 
 | |
