8F6R
 
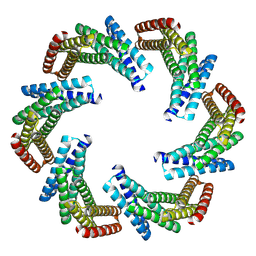 | | CryoEM structure of designed modular protein oligomer C6-79 | | Descriptor: | De novo designed oligomeric protein C6-79 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Cell, 2024
|
|
8F6Q
 
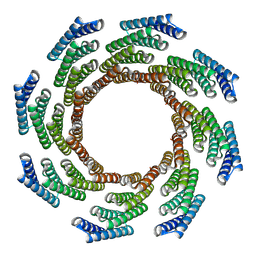 | | CryoEM structure of designed modular protein oligomer C8-71 | | Descriptor: | C8-71 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Biorxiv, 2023
|
|
6G7K
 
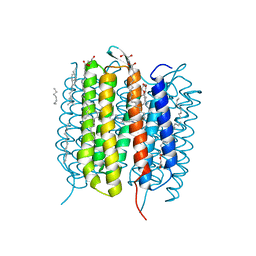 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 10 ps state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
1HLW
 
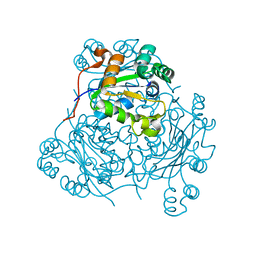 | | STRUCTURE OF THE H122A MUTANT OF THE NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Admiraal, S.J, Meyer, P, Schneider, B, Deville-Bonne, D, Janin, J, Herschlag, D. | | Deposit date: | 2000-12-04 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical rescue of phosphoryl transfer in a cavity mutant: a cautionary tale for site-directed mutagenesis.
Biochemistry, 40, 2001
|
|
2EXI
 
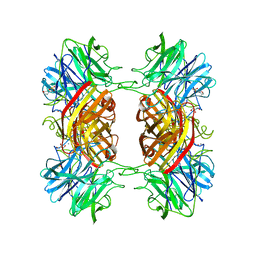 | | Structure of the family43 beta-Xylosidase D15G mutant from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2ERL
 
 | | PHEROMONE ER-1 FROM | | Descriptor: | ETHANOL, MATING PHEROMONE ER-1 | | Authors: | Anderson, D.H, Weiss, M.S, Eisenberg, D. | | Deposit date: | 1995-12-20 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1OXQ
 
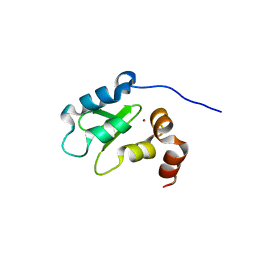 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AVPIAQKSE (Smac) peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
1HK6
 
 | | Ral binding domain from Sec5 | | Descriptor: | EXOCYST COMPLEX COMPONENT SEC5 | | Authors: | Mott, H.R, Nietlispach, D, Hopkins, L.J, Mirey, G, Camonis, J.H, Owen, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the GTPase-binding domain of Sec5 and elucidation of its Ral binding site.
J. Biol. Chem., 278, 2003
|
|
2CJA
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-30 | | Release date: | 2006-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2C8O
 
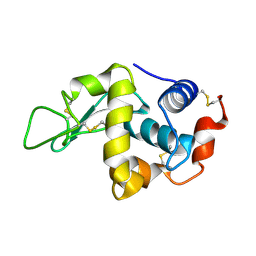 | | lysozyme (1sec) and UV lasr excited fluorescence | | Descriptor: | LYSOZYME C | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3BPV
 
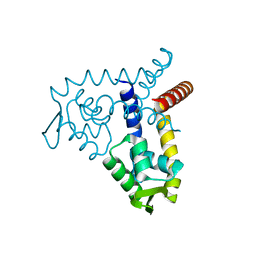 | | Crystal Structure of MarR | | Descriptor: | Transcriptional regulator | | Authors: | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
2CIM
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2EZ1
 
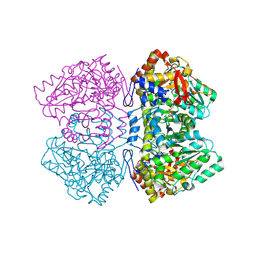 | | Holo tyrosine phenol-lyase from Citrobacter freundii at pH 8.0 | | Descriptor: | POTASSIUM ION, Tyrosine phenol-lyase | | Authors: | Milic, D, Matkovic-Calogovic, D, Demidkina, T.V, Antson, A.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-07-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of apo- and holo-tyrosine phenol-lyase reveal a catalytically critical closed conformation and suggest a mechanism for activation by K+ ions
Biochemistry, 45, 2006
|
|
2F8U
 
 | | G-quadruplex structure formed in human Bcl-2 promoter, hybrid form | | Descriptor: | 5'-D(*GP*GP*GP*CP*GP*CP*GP*GP*GP*AP*GP*GP*AP*AP*TP*TP*GP*GP*GP*CP*GP*GP*G)-3' | | Authors: | Dai, J, Chen, D, Carver, M, Yang, D. | | Deposit date: | 2005-12-03 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region.
Nucleic Acids Res., 34, 2006
|
|
3AG9
 
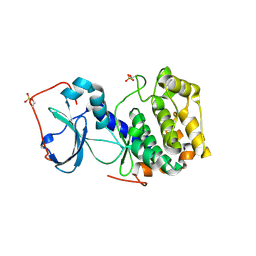 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1012 | | Descriptor: | (10R,20R,23R)-10-(4-aminobutyl)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamidopropyl)-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Lavogina, D, Uri, A, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2010-03-26 | | Release date: | 2010-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
2FLD
 
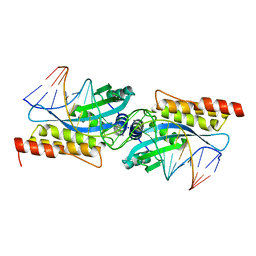 | | I-MsoI Re-Designed for Altered DNA Cleavage Specificity | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Ashworth, J, Duarte, C.M, Havranek, J.J, Sussman, D, Monnat, R.J, Stoddard, B.L, Baker, D. | | Deposit date: | 2006-01-05 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational redesign of endonuclease DNA binding and cleavage specificity.
Nature, 441, 2006
|
|
1LJM
 
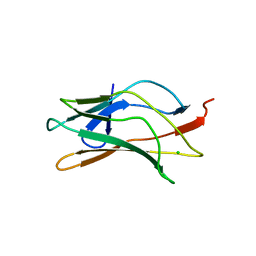 | | DNA recognition is mediated by conformational transition and by DNA bending | | Descriptor: | CHLORIDE ION, RUNX1 transcription factor | | Authors: | Bartfeld, D, Shimon, L, Couture, G.C, Rabinovich, D, Frolow, F, Levanon, D, Groner, Y, Shakked, Z. | | Deposit date: | 2002-04-22 | | Release date: | 2002-11-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA Recognition by the RUNX1 Transcription Factor Is Mediated by an Allosteric Transition in the RUNT Domain and by DNA Bending.
Structure, 10
|
|
2EXK
 
 | | Structure of the family43 beta-Xylosidase E187G from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2CJB
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with serine | | Descriptor: | CHLORIDE ION, SERINE, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-30 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
3BD9
 
 | | human 3-O-sulfotransferase isoform 5 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Xu, D, Moon, A.F, Song, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering sulfotransferases to modify heparan sulfate.
Nat.Chem.Biol., 4, 2008
|
|
2EXH
 
 | | Structure of the family43 beta-Xylosidase from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Yuval, S, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EZ2
 
 | | Apo tyrosine phenol-lyase from Citrobacter freundii at pH 8.0 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Tyrosine phenol-lyase | | Authors: | Milic, D, Matkovic-Calogovic, D, Demidkina, T.V, Antson, A.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of apo- and holo-tyrosine phenol-lyase reveal a catalytically critical closed conformation and suggest a mechanism for activation by K+ ions
Biochemistry, 45, 2006
|
|
2FOF
 
 | | Structure of porcine pancreatic elastase in 80% isopropanol | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2FOB
 
 | | Structure of porcine pancreatic elastase in 40/50/10 cyclohexane | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2FOA
 
 | | Structure of porcine pancreatic elastase in 40/50/10 % benzene | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
