2O4X
 
 | | Crystal structure of human P100 tudor domain | | Descriptor: | Staphylococcal nuclease domain-containing protein 1 | | Authors: | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human P100 tudor domain
To be Published
|
|
2QVO
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-08-08 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of AF1382 from Archaeoglobus fulgidus.
To be Published
|
|
2PG0
 
 | | Crystal structure of acyl-CoA dehydrogenase from Geobacillus kaustophilus | | Descriptor: | Acyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Chen, L, Chen, L.-Q, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Li, Y, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of acyl-CoA dehydrogenase from G. kaustophilus
To be Published
|
|
2P8T
 
 | | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH0730 | | Authors: | Chen, L, Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Fu, Z.-Q, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3
To be Published
|
|
2P68
 
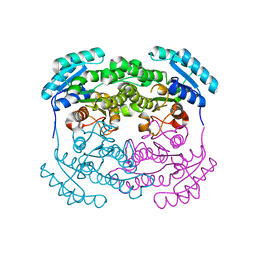 | | Crystal Structure of aq_1716 from Aquifex Aeolicus VF5 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Chen, L, Chen, L.-Q, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Dillard, B, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of aq_1716 from Aquifex aeolicus VF5
To be Published
|
|
7FB3
 
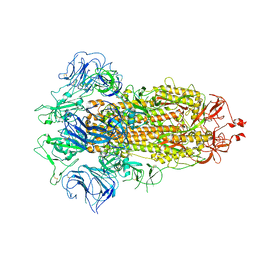 | |
7FB0
 
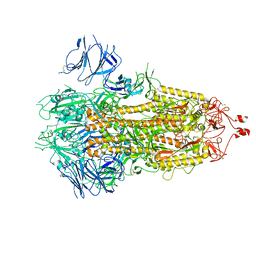 | | SARS-CoV-2 spike protein in closed state | | Descriptor: | Spike glycoprotein | | Authors: | Zhu, Y, Tai, L.H, Sun, F. | | Deposit date: | 2021-07-08 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Novel sites for Cathepsin L cleavage in SARS-CoV-2 spike guide treatment strategies
To Be Published
|
|
7FB1
 
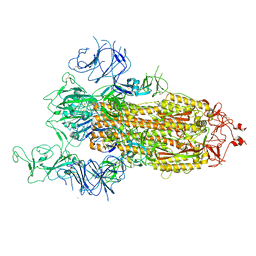 | |
7FB4
 
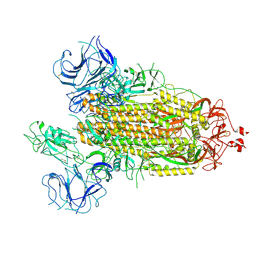 | |
5Z1P
 
 | |
5GSV
 
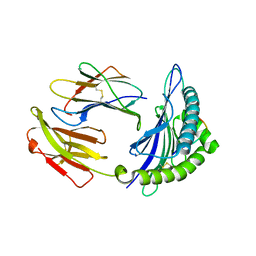 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-5 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GR7
 
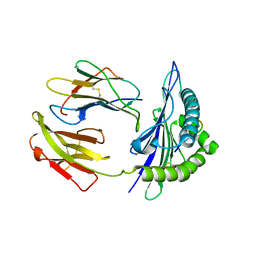 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSR
 
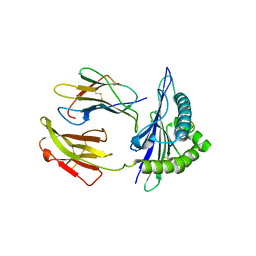 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | Descriptor: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSB
 
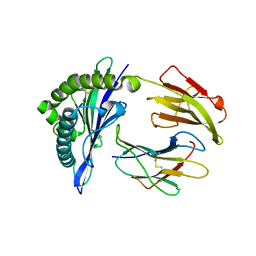 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-3 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
3SGM
 
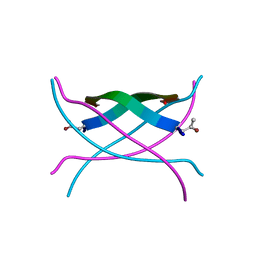 | | Bromoderivative-2 of amyloid-related segment of alphaB-crystallin residues 90-100 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin B chain | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7006 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
3SGR
 
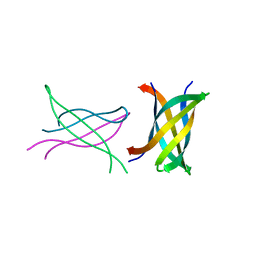 | | Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
3SGN
 
 | |
3SGO
 
 | |
3SGS
 
 | |
3SGP
 
 | | Amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin B chain | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4016 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
5WVW
 
 | | The crystal structure of Cren7 mutant L28A in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WWC
 
 | | The crystal structure of Cren7 mutant L28M in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-31 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVZ
 
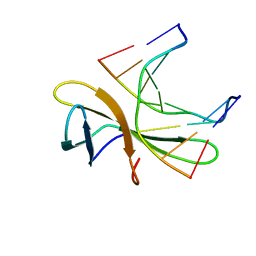 | | The crystal structure of Cren7 mutant L28F in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-30 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVY
 
 | | The crystal structure of Cren7 mutant L28V in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
2Y29
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph III | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
