7DCB
 
 | |
7DCW
 
 | |
7DM6
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase complexed with crotonoside | | Descriptor: | 6-azanyl-9-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1H-purin-2-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DQN
 
 | |
7DH1
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N2-Methylguanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-12 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DGC
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with 2'-O-Methylguanosine | | Descriptor: | 9-[(2R,3R,4R)-5-(hydroxymethyl)-3-methoxy-4-oxidanyl-oxolan-2-yl]-3H-purine-2,6-dione, Guanosine deaminase, SODIUM ION, ... | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DM5
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with guanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DPK
 
 | |
7DLC
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N1-methylguanosine | | Descriptor: | 2-azanyl-9-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1-methyl-purin-6-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7DOX
 
 | |
7DOW
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with 7-deazaguansoine | | Descriptor: | 2-azanyl-7-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-3H-pyrrolo[2,3-d]pyrimidin-4-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7DOY
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in complex with 6-O-methylguanosine | | Descriptor: | (2R,3R,4S,5R)-2-(2-azanyl-6-methoxy-purin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7E8L
 
 | | The structure of Spodoptera litura chemosensory protein | | Descriptor: | Putative chemosensory protein CSP8 | | Authors: | Xie, W, Jia, Q, Zeng, H, Xiao, N, Tang, J, Gao, S, Zhang, J. | | Deposit date: | 2021-03-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Spodoptera litura Chemosensory Protein CSP8.
Insects, 12, 2021
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR5
 
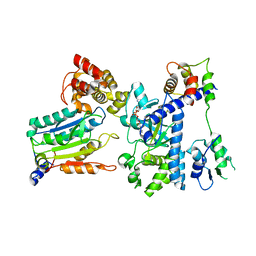 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | Descriptor: | Calmodulin-1, Caspase-4, OspC3, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR4
 
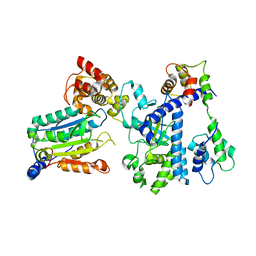 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | Descriptor: | Calmodulin-1, Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR0
 
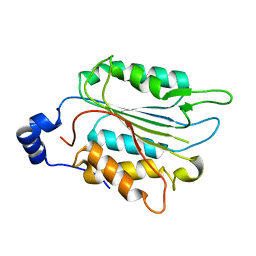 | | P32 of caspase-4 C258A mutant | | Descriptor: | Caspase-4 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR6
 
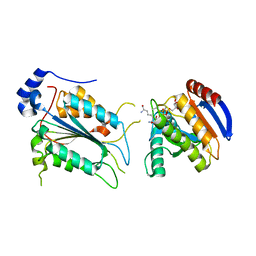 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | Descriptor: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR1
 
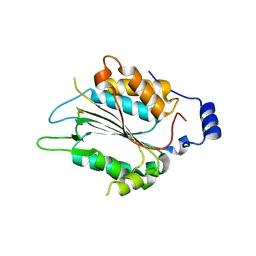 | | P32 of caspase-4 C258A mutant in complex with OspC3 C-terminal ankyrin-repeat domain | | Descriptor: | Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR2
 
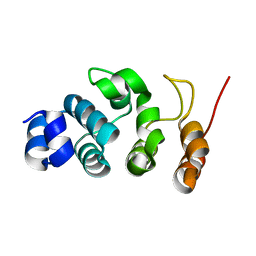 | |
3NCW
 
 | |
1DPR
 
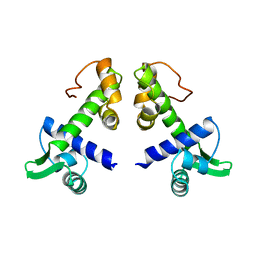 | | STRUCTURES OF THE APO-AND METAL ION ACTIVATED FORMS OF THE DIPHTHERIA TOX REPRESSOR FROM CORYNEBACTERIUM DIPHTHERIAE | | Descriptor: | DIPHTHERIA TOX REPRESSOR | | Authors: | Schiering, N, Tao, X, Murphy, J, Petsko, G.A, Ringe, D. | | Deposit date: | 1995-02-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the apo- and the metal ion-activated forms of the diphtheria tox repressor from Corynebacterium diphtheriae.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
6KMU
 
 | | P22/P10 complex of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KMZ
 
 | |
6KMT
 
 | | P32 of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
