8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
6WWB
 
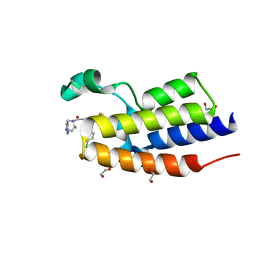 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the compound 3b | | Descriptor: | 1,2-ETHANEDIOL, 2-((S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)-N-((1-(4-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)acetamido)butyl)-1H-1,2,3-triazol-4-yl)methyl)acetamide, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Phenyl-Glutarimides: Alternative Cereblon Binders for the Design of PROTACs.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4NIE
 
 | |
9IM5
 
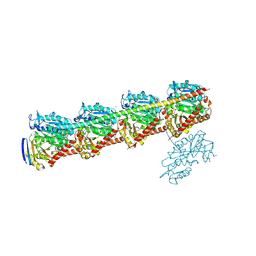 | | Tubulin-RB3(MUT)-TTL-Y12 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-02 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IMO
 
 | | Crystal structure of Tubulin-RB3-TTL-Y12 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-04 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7B7V
 
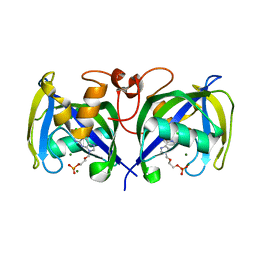 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
6XER
 
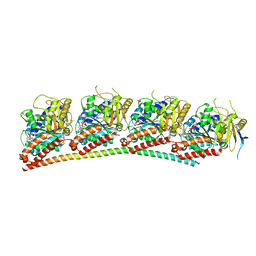 | | Tubulin-RB3_SLD in complex with colchicine | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-06-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Stable Colchicine-Binding Site Tubulin Inhibitors 6-Aryl-2-benzoyl-pyridines as Potential Anticancer Agents.
J.Med.Chem., 64, 2021
|
|
6XET
 
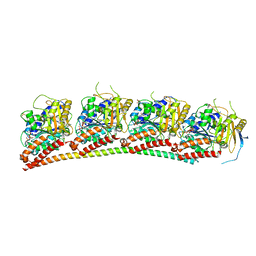 | | Tubulin-RB3_SLD in complex with compound 60c | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-06-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Stable Colchicine-Binding Site Tubulin Inhibitors 6-Aryl-2-benzoyl-pyridines as Potential Anticancer Agents.
J.Med.Chem., 64, 2021
|
|
6XES
 
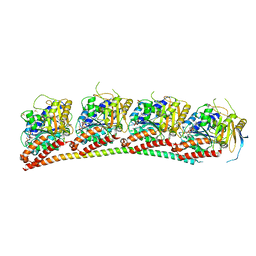 | | Tubulin-RB3_SLD in complex with compound 40a | | Descriptor: | DIMETHYL SULFOXIDE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-06-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Stable Colchicine-Binding Site Tubulin Inhibitors 6-Aryl-2-benzoyl-pyridines as Potential Anticancer Agents.
J.Med.Chem., 64, 2021
|
|
7LZE
 
 | | Cryo-EM Structure of disulfide stabilized HMPV F v4-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8DZI
 
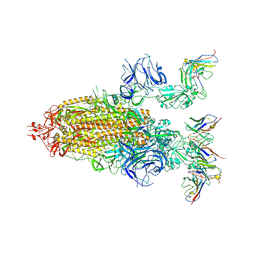 | |
6PKA
 
 | |
8DZH
 
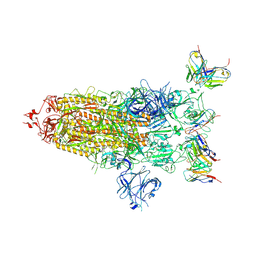 | |
6PMD
 
 | |
9MS1
 
 | | Crystal structure of human CYP3A4 in complex with SJYHJ-111 | | Descriptor: | Cytochrome P450 3A4, N'-(2-chlorophenyl)-N-[(3-hydroxyphenyl)methyl]-N-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]urea, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
9MS2
 
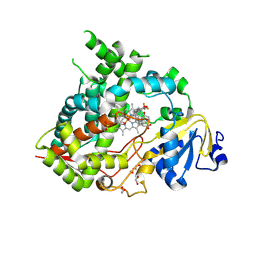 | | Crystal structure of human CYP3A5 in complex with SJYHJ-111 | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 3A5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
6J20
 
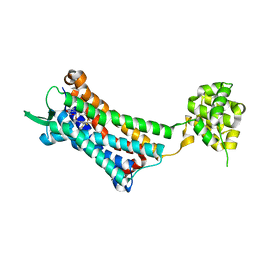 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
6J21
 
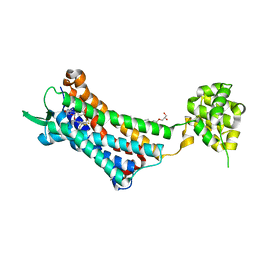 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
9IQE
 
 | |
9IQF
 
 | |
9IQG
 
 | | Cryo-EM structure of MsRv1273c/72c from Mycobacterium smegmatis in the ATP|ADP+Vi-bound Occ (Vi) state | | Descriptor: | ABC transporter transmembrane region, ABC transporter, ATP-binding protein, ... | | Authors: | Lan, Y, Yu, J, Li, J. | | Deposit date: | 2024-07-12 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of a mycobacterial isoniazid efflux pump MsRv1273c/72c with a degenerate nucleotide-binding site.
Nat Commun, 16, 2025
|
|
7PAD
 
 | | The crystal structure of DW-0254 in complex with PDE6D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
7PAC
 
 | | The crystal structure of PDE6D in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
