1VPZ
 
 | |
1VKH
 
 | |
1EI3
 
 | | CRYSTAL STRUCTURE OF NATIVE CHICKEN FIBRINOGEN | | Descriptor: | FIBRINOGEN | | Authors: | Yang, Z, Mochalkin, I, Veerapandian, L, Riley, M, Doolittle, R.F. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal structure of native chicken fibrinogen at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2ETS
 
 | |
2HIW
 
 | |
2FNO
 
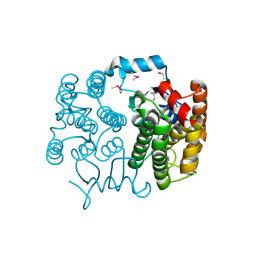 | |
1B8K
 
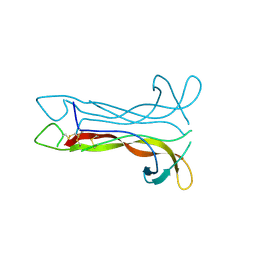 | | Neurotrophin-3 from Human | | Descriptor: | PROTEIN (NEUROTROPHIN-3) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1B8M
 
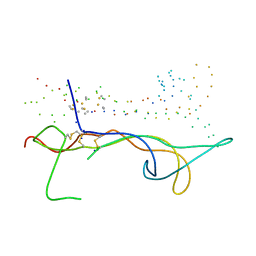 | | BRAIN DERIVED NEUROTROPHIC FACTOR, NEUROTROPHIN-4 | | Descriptor: | PROTEIN (BRAIN DERIVED NEUROTROPHIC FACTOR), PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
2HUJ
 
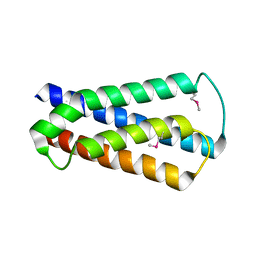 | |
1N86
 
 | | Crystal structure of human D-dimer from cross-linked fibrin complexed with GPR and GHRPLDK peptide ligands. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, Fibrin alpha/alpha-E chain, ... | | Authors: | Yang, Z, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-11-19 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of fragment double-D from cross-linked lamprey fibrin reveals isopeptide linkages across an unexpected D-D interface.
Biochemistry, 41, 2002
|
|
1M1J
 
 | | Crystal structure of native chicken fibrinogen with two different bound ligands | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, Z, Kollman, J.M, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Native Chicken Fibrinogen at 2.7 A Resolution
Biochemistry, 40, 2001
|
|
3O0P
 
 | | Pilus-related Sortase C of Group B Streptococcus | | Descriptor: | Sortase family protein | | Authors: | Malito, E, Spraggon, G. | | Deposit date: | 2010-07-19 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus.
Faseb J., 25, 2011
|
|
1J5Y
 
 | |
1J5S
 
 | |
3LCT
 
 | |
3L9P
 
 | |
1O58
 
 | |
6TCZ
 
 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 | | Descriptor: | Proteasome endopeptidase complex, Proteasome subunit alpha type, Proteasome subunit beta, ... | | Authors: | Srinivas, H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
3LCS
 
 | |
6TD5
 
 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | Authors: | Srinivas, H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-08-26 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
1O5U
 
 | |
1O4W
 
 | |
1O2D
 
 | |
1O4U
 
 | |
1O1X
 
 | |
