8QBF
 
 | | Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOSERINE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 2024
|
|
8QB7
 
 | |
8QBD
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10) | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 2024
|
|
8QBG
 
 | |
8QBE
 
 | |
8R61
 
 | |
8RZE
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 10 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-pyridin-3-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 10
To Be Published
|
|
8RZC
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-imidazol-1-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11
To Be Published
|
|
8RZD
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 9 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 9
To Be Published
|
|
8RUU
 
 | | Fabs derived from bimekizumab in complex with IL-17F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoblobulin heavy chain, Immunoblobulin light chain, ... | | Authors: | Adams, R, Lawson, A.D.G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Bimekizumab Fab Fragment in Complex with IL-17F Provides Molecular Basis for Dual IL-17A and IL-17F Inhibition.
J Invest Dermatol., 2024
|
|
8QZO
 
 | |
8S8A
 
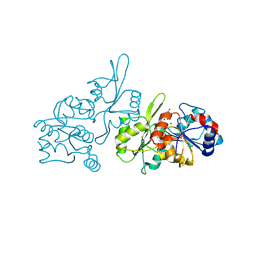 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone without phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, CHLORIDE ION, Chronophin, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
8R15
 
 | | Crystal structure of Fusarium oxysporum NADase I | | Descriptor: | GLYCEROL, TNT domain-containing protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of fungal tuberculosis necrotizing toxin (TNT) domain-containing enzymes reveals divergent adaptations to enhance NAD cleavage.
Protein Sci., 33, 2024
|
|
8SP7
 
 | |
8SO0
 
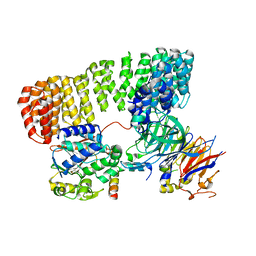 | | Cryo-EM structure of the PP2A:B55-FAM122A complex | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-04-28 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.79961 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
8QY4
 
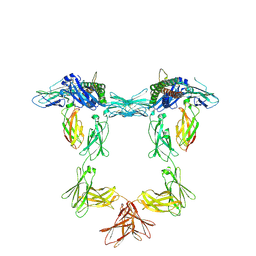 | | Structure of interleukin 11 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure of interleukin 11 (gp130 P496L mutant).
To Be Published
|
|
8QY6
 
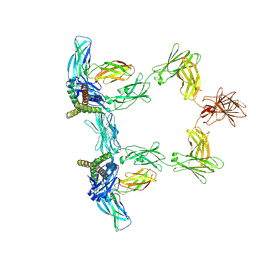 | | Structure of interleukin 6 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of interleukin 6 (gp130 P496L mutant).
To Be Published
|
|
8QY5
 
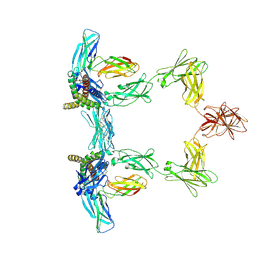 | | Structure of interleukin 6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of interleukin 6.
To Be Published
|
|
8RIV
 
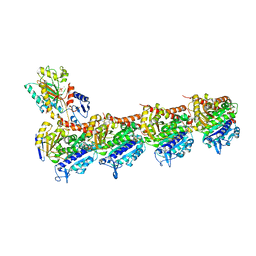 | | T2R-TTL-1-K08 complex | | Descriptor: | (4-fluoranyl-2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E, Prota, A.E.P. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8RIW
 
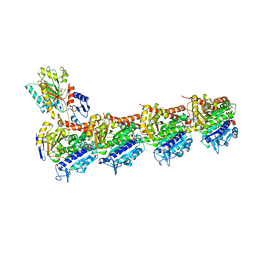 | | T2R-TTL-1-L01 complex | | Descriptor: | (2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E.P, Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8QU4
 
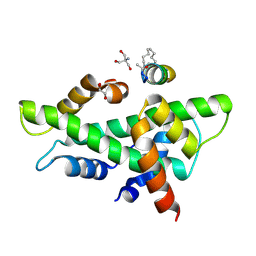 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker in an alternative binding pose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU3
 
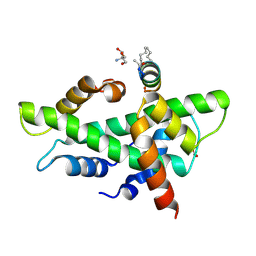 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU2
 
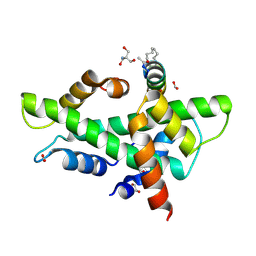 | | NF-YB/C Heterodimer in Complex with a 16-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Durukan, C, Arbore, F, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8ROO
 
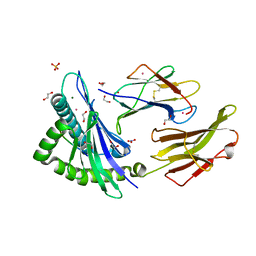 | | Crystal structure of HLA B*18:01 in complex with YERMCNIL, an 8-mer epitope from Influenza A | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Influenza A derived peptide/Nucleoprotein, ... | | Authors: | Murdolo, L.D, Maddumage, J.C, Gras, S. | | Deposit date: | 2024-01-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterisation of novel influenza-derived HLA-B*18:01-restricted epitopes.
Clin Transl Immunology, 13, 2024
|
|
8ROP
 
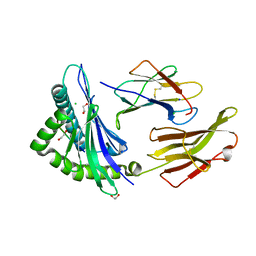 | | Crystal structure of HLA B*18:01 in complex with QEIRTFSF, an 8-mer epitope from Influenza A | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Murdolo, L.D, Maddumage, J.C, Gras, S. | | Deposit date: | 2024-01-12 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Characterisation of novel influenza-derived HLA-B*18:01-restricted epitopes.
Clin Transl Immunology, 13, 2024
|
|
