6UQU
 
 | |
6UR3
 
 | |
6VXG
 
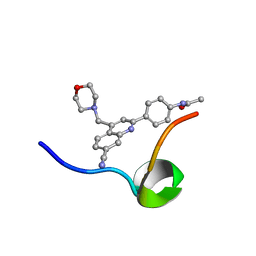 | | Structure of the C-terminal Domain of RAGE and Its Inhibitor | | Descriptor: | Advanced glycosylation end product-specific receptor, N-(4-{7-cyano-4-[(morpholin-4-yl)methyl]quinolin-2-yl}phenyl)acetamide | | Authors: | Ramirez, L, Shekhtman, A. | | Deposit date: | 2020-02-21 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small-molecule antagonism of the interaction of the RAGE cytoplasmic domain with DIAPH1 reduces diabetic complications in mice.
Sci Transl Med, 13, 2021
|
|
3CIB
 
 | | Structure of BACE Bound to SCH727596 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4S)-4-benzylpiperidin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4XUK
 
 | |
6IUI
 
 | | Crystal structure of GIT1 PBD domain in complex with Paxillin LD4 motif | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the target-binding mode of the G protein-coupled receptor kinase-interacting protein in the regulation of focal adhesion dynamics.
J. Biol. Chem., 294, 2019
|
|
6IUH
 
 | | Crystal structure of GIT1 PBD domain in complex with Liprin-alpha2 | | Descriptor: | ARF GTPase-activating protein GIT1, IODIDE ION, Liprin-alpha-2 | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the target-binding mode of the G protein-coupled receptor kinase-interacting protein in the regulation of focal adhesion dynamics.
J. Biol. Chem., 294, 2019
|
|
7UP2
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP1
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP3
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (3P)-4-[4-(hydroxymethyl)phenyl]-3-(2H-tetrazol-5-yl)pyridine-2-sulfonamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOX
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-(hydroxymethyl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-ol, ACETATE ION, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOY
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (6P)-4-amino-6-(2H-tetrazol-5-yl)benzene-1,3-disulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
6LAN
 
 | | Structure of CCDC50 and LC3B complex | | Descriptor: | Coiled-coil domain-containing protein 50,Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Liu, L, Li, J, Hou, P. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection.
Cell Res., 31, 2021
|
|
4IRE
 
 | | Crystal structure of GLIC with mutations at the loop C region | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, OXALATE ION, ... | | Authors: | Chen, Q, Pan, J, Liang, Y.H, Xu, Y, Tang, P. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Signal transduction pathways in the pentameric ligand-gated ion channels.
Plos One, 8, 2013
|
|
7T7A
 
 | | Crystal Structure of Human SHOC2: A Leucine-Rich Repeat Protein | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, NITRATE ION | | Authors: | Hajian, B, Lemke, C, Kwon, J, Bian, Y, Fuller, C, Aguirre, J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure-function analysis of the SHOC2-MRAS-PP1C holophosphatase complex.
Nature, 609, 2022
|
|
6KN9
 
 | |
4X1K
 
 | | Discovery of cytotoxic Dolastatin 10 analogs with N-terminal modifications | | Descriptor: | 2-methyl-L-alanyl-N-[(3R,4S,5S)-1-{(2S)-2-[(1R,2R)-3-{[(1S)-1-carboxy-2-phenylethyl]amino}-1-methoxy-2-methyl-3-oxopropyl]pyrrolidin-1-yl}-3-methoxy-5-methyl-1-oxoheptan-4-yl]-N-methyl-L-valinamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parris, K.D. | | Deposit date: | 2014-11-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of cytotoxic dolastatin 10 analogues with N-terminal modifications.
J.Med.Chem., 57, 2014
|
|
7UYB
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)-4-(trifluoromethyl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYC
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2P)-4'-(piperidin-4-yl)-4-[(piperidin-4-yl)methyl]-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYA
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYD
 
 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
4YPI
 
 | | Structure of Ebola virus nucleoprotein N-terminal fragment bound to a peptide derived from Ebola VP35 | | Descriptor: | Nucleoprotein, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Borek, D.M, Binning, J.M, Otwinowski, Z, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-08 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions.
Cell Rep, 11, 2015
|
|
3J24
 
 | | CryoEM reconstruction of complement decay-accelerating factor | | Descriptor: | Complement decay-accelerating factor | | Authors: | Yoder, J.D, Hafenstein, S.H. | | Deposit date: | 2012-08-17 | | Release date: | 2012-09-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | The Crystal Structure of a Coxsackievirus B3-RD Variant and a Refined 9-Angstrom Cryo-Electron Microscopy Reconstruction of the Virus Complexed with Decay-Accelerating Factor (DAF) Provide a New Footprint of DAF on the Virus Surface.
J.Virol., 86, 2012
|
|
2R14
 
 | | Structure of morphinone reductase in complex with tetrahydroNAD | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, Morphinone reductase | | Authors: | Costello, C.L, Scrutton, N.S, Leys, D. | | Deposit date: | 2007-08-22 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenesis of morphinone reductase induces multiple reactive configurations and identifies potential ambiguity in kinetic analysis of enzyme tunneling mechanisms.
J.Am.Chem.Soc., 129, 2007
|
|
6MIS
 
 | | Native ananain in complex with E-64 | | Descriptor: | Ananain, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Yongqing, T, Wilmann, P.G, Pike, R.N, Wijeyewickrema, L.C. | | Deposit date: | 2018-09-20 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Determination of the crystal structure and substrate specificity of ananain.
Biochimie, 166, 2019
|
|
