7E8G
 
 | |
7E89
 
 | |
7E8B
 
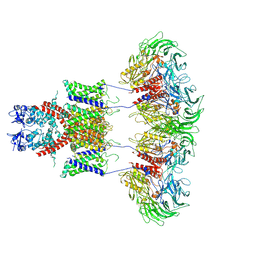 | | CryoEM structure of human Kv4.2-DPP6S complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E8H
 
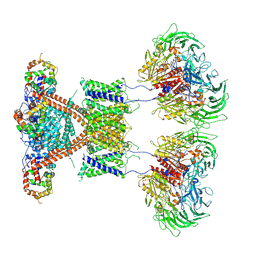 | | CryoEM structure of human Kv4.2-DPP6S-KChIP1 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7F0J
 
 | | CryoEM structure of human Kv4.2 | | Descriptor: | Potassium voltage-gated channel subfamily D member 2, ZINC ION | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-06-04 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7F3F
 
 | | CryoEM structure of human Kv4.2-KChIP1 complex | | Descriptor: | Isoform 2 of Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7VSG
 
 | |
7VSH
 
 | |
7EFL
 
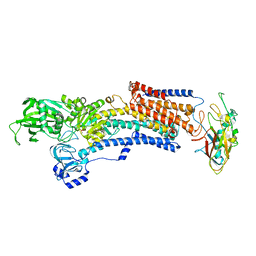 | | Crystal structure of the gastric proton pump K791S in (BYK)E2BeF state | | Descriptor: | (7R,8R,9S)-2,3-dimethyl-9-phenyl-7,8,9,10-tetrahydroimidazo[1,2-h][1,7]naphthyridine-7,8-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Yamamoto, K, Irie, K. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Gastric proton pump with two occluded K + engineered with sodium pump-mimetic mutations.
Nat Commun, 12, 2021
|
|
7EFM
 
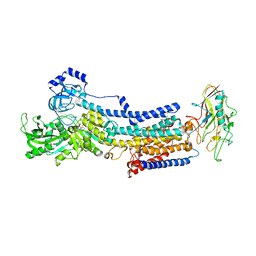 | | Crystal structure of the gastric proton pump K791S/E820D/Y340N in (BYK)E2BeF state | | Descriptor: | (7R,8R,9S)-2,3-dimethyl-9-phenyl-7,8,9,10-tetrahydroimidazo[1,2-h][1,7]naphthyridine-7,8-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Yamamoto, K, Irie, K. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gastric proton pump with two occluded K + engineered with sodium pump-mimetic mutations.
Nat Commun, 12, 2021
|
|
7EFN
 
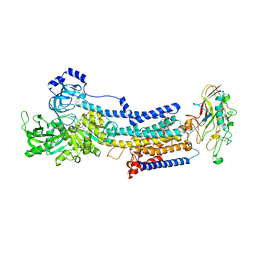 | | Crystal structure of the gastric proton pump K791S/E820D/Y340N/E936V in (BYK)E2BeF state | | Descriptor: | (7R,8R,9S)-2,3-dimethyl-9-phenyl-7,8,9,10-tetrahydroimidazo[1,2-h][1,7]naphthyridine-7,8-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Yamamoto, K, Irie, K. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gastric proton pump with two occluded K + engineered with sodium pump-mimetic mutations.
Nat Commun, 12, 2021
|
|
7ET1
 
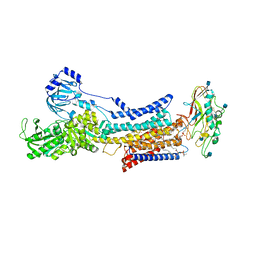 | |
7XJH
 
 | | Isoproterenol-activated dog beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the beta 3-adrenergic receptor reveals the molecular basis of subtype selectivity.
Mol.Cell, 81, 2021
|
|
7X22
 
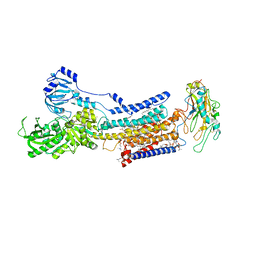 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794S in (2K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Nakanishi, H, Abe, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
7X20
 
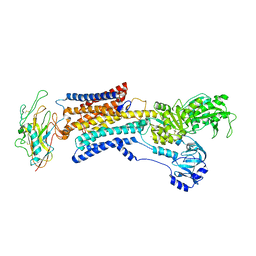 | |
7X24
 
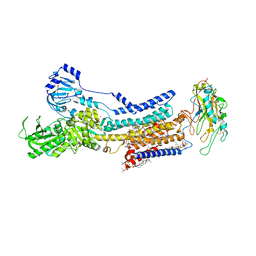 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 SPWC mutant in (2K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Nakanishi, H, Young, V, Artigas, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
7X21
 
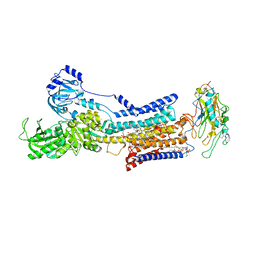 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794A in (K+)E2-AlF state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Nakanishi, H, Abe, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
7X23
 
 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 SPWC mutant in 3Na+E1-AMPPCPF state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase alpha chain 2, ... | | Authors: | Abe, K, Nakanishi, H, Young, V, Artigas, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
7F47
 
 | | Cryo-EM structure of Rhizobium etli MprF | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Hypothetical conserved protein, [(2R)-1-[[(2R)-3-[(2S)-2,6-bis(azanyl)hexanoyl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (E)-octadec-9-enoate | | Authors: | Nishimura, M, Hirano, H, Kobayashi, K, Gill, C.P, Phan, C.N.K, Kise, Y, Kusakizako, T, Yamashita, K, Ito, Y, Roy, H, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of Rhizobium etli MprF
To Be Published
|
|
7EFC
 
 | | 1.70 A cryo-EM structure of streptavidin | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | 1.70 A cryo-EM structure of streptavidin using all frames (corresponding to 70 e/A^2 total dose)
To Be Published
|
|
7EFD
 
 | | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40 e/A^2 total dose) | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.77 Å) | | Cite: | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40e/A^2 total dose)
To Be Published
|
|
7DY0
 
 | |
