1SLN
 
 | |
6Z0F
 
 | | Crystal structure of the membrane pseudokinase YukC/EssB from Bacillus subtilis T7SS | | Descriptor: | ESX secretion system protein YukC | | Authors: | Tassinari, M, Bellinzoni, M, Alzari, P.M, Fronzes, R, Gubellini, F. | | Deposit date: | 2020-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Antibacterial Type VII Secretion System of Bacillus subtilis: Structure and Interactions of the Pseudokinase YukC/EssB.
Mbio, 13, 2022
|
|
4FBN
 
 | | Insights into structural integration of the PLCgamma regulatory region and mechanism of autoinhibition and activation based on key roles of SH2 domains | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
8QOE
 
 | | Inward-facing conformation of the ABC transporter BmrA | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Di Cesare, M, Kaplan, E, Valimehr, S, Hanssen, E, Orelle, C, Jault, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.
J.Biol.Chem., 300, 2023
|
|
6I8F
 
 | |
6RB0
 
 | |
6RKY
 
 | |
2HDP
 
 | | Solution Structure of Hdm2 RING Finger Domain | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2, ZINC ION | | Authors: | Kostic, M, Matt, T, Yamout-Martinez, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2006-06-20 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53.
J.Mol.Biol., 363, 2006
|
|
3ECW
 
 | |
3ECV
 
 | |
3ECU
 
 | |
5K4I
 
 | | Crystal Structure of ERK2 in complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
5K4J
 
 | | Crystal Structure of CDK2 in complex with compound 22 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Cyclin-dependent kinase 2 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
8DXT
 
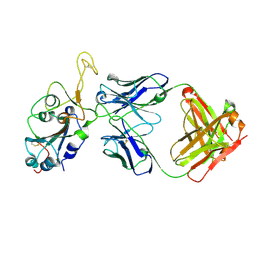 | | Fab arm of antibody GAR12 bound to the receptor binding domain of SARS-CoV-2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab arm of antibody GAR12, Light chain of Fab arm of antibody GAR12, ... | | Authors: | Langley, D.B, Christ, D, Henry, J.Y. | | Deposit date: | 2022-08-03 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Nat Commun, 14, 2023
|
|
8DXU
 
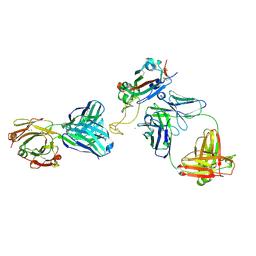 | |
8CHB
 
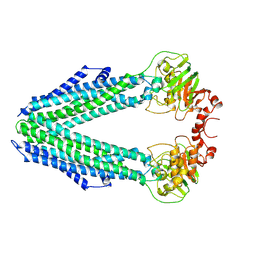 | | Inward-facing conformation of the ABC transporter BmrA C436S/A582C cross-linked mutant | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Di Cesare, M, Kaplan, E, Hanssen, E, Valimehr, S, Orelle, C, Jault, J.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.
J.Biol.Chem., 300, 2023
|
|
5NCR
 
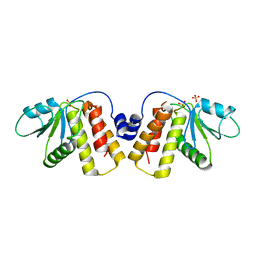 | | OH1 from the Orf virus: a tyrosine phosphatase that displays distinct structural features and triple substrate specificity | | Descriptor: | PHOSPHATE ION, SULFATE ION, tyrosine phosphatase | | Authors: | Segovia, D, Haouz, A, Berois, M, Villarino, A, Andre-Leroux, G. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | OH1 from Orf Virus: A New Tyrosine Phosphatase that Displays Distinct Structural Features and Triple Substrate Specificity.
J. Mol. Biol., 429, 2017
|
|
6SCP
 
 | |
6SAT
 
 | |
6SCQ
 
 | |
6SCS
 
 | |
3D4Q
 
 | | Pyrazole-based inhibitors of B-Raf kinase | | Descriptor: | (1E)-5-(1-piperidin-4-yl-3-pyridin-4-yl-1H-pyrazol-4-yl)-2,3-dihydro-1H-inden-1-one oxime, B-Raf proto-oncogene serine/threonine-protein kinase | | Authors: | Morales, T, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2008-05-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent and selective pyrazole-based inhibitors of B-Raf kinase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4XJ0
 
 | | Crystal structure of ERK2 in complex with an inhibitor 14K | | Descriptor: | 1-[(1S)-1-(4-chloro-3-fluorophenyl)-2-hydroxyethyl]-4-[2-(tetrahydro-2H-pyran-4-ylamino)pyrimidin-4-yl]pyridin-2(1H)-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2015-01-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of highly potent, selective, and efficacious small molecule inhibitors of ERK1/2.
J.Med.Chem., 58, 2015
|
|
6QPL
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor MS31 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Johansson, C, Krojer, T, Xiong, Y, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Potent and Selective Fragment-like Inhibitor of Methyllysine Reader Protein Spindlin 1 (SPIN1).
J.Med.Chem., 62, 2019
|
|
3PSB
 
 | | Furo[2,3-c]pyridine-based Indanone Oximes as Potent and Selective B-Raf Inhibitors | | Descriptor: | B-RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE, ethyl 3-{[1-(hydroxyamino)-2H-inden-5-yl]amino}thieno[2,3-c]pyridine-2-carboxylate | | Authors: | Morales, T, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-12-01 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Discovery of furo[2,3-c]pyridine-based indanone oximes as potent and selective B-Raf inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
