1WOI
 
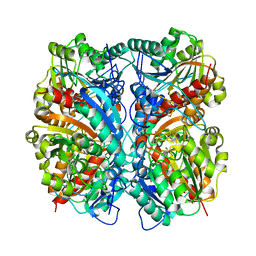 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
3TUC
 
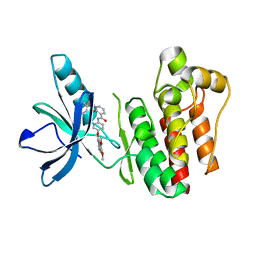 | | Crystal structure of SYK kinase domain with 1-benzyl-N-(5-(6,7-dimethoxyquinolin-4-yloxy)pyridin-2-yl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Descriptor: | 1-benzyl-N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-2-oxo-1,2-dihydropyridine-3-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
3TUD
 
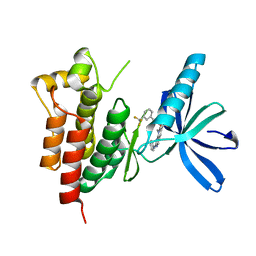 | | Crystal structure of SYK kinase domain with N-(4-methyl-3-(8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl)phenyl)-3-(trifluoromethyl)benzamide | | Descriptor: | N-{4-methyl-3-[8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl]phenyl}-3-(trifluoromethyl)benzamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
5F5R
 
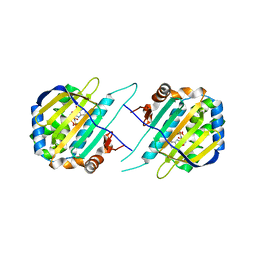 | | TRAP1N-ADPNP | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, MAGNESIUM ION, ... | | Authors: | Tsai, F.T.F, Lee, S, Sung, N, Lee, J, Chang, C, Joachimiak, A. | | Deposit date: | 2015-12-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4MN0
 
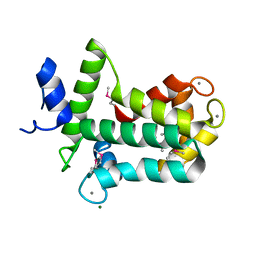 | | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca2+-loaded apoprotein conformation state | | Descriptor: | Berovin, CALCIUM ION, MAGNESIUM ION | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca(2+)-loaded apoprotein conformation state.
Biochim.Biophys.Acta, 1834, 2013
|
|
5YO9
 
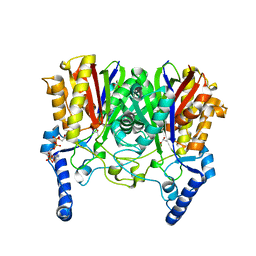 | |
5YOA
 
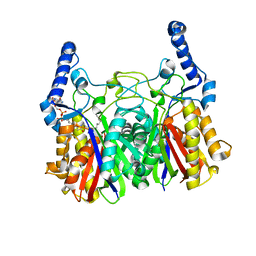 | |
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
3N2O
 
 | | X-ray crystal structure of arginine decarboxylase complexed with Arginine from Vibrio vulnificus | | Descriptor: | AGMATINE, Biosynthetic arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
4N1F
 
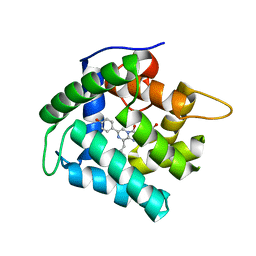 | | Crystal Structure of F88Y obelin mutant from Obelia longissima at 2.09 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Natashin, P.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-10-04 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Crystal structures of the F88Y obelin mutant before and after bioluminescence provide molecular insight into spectral tuning among hydromedusan photoproteins
Febs J., 281, 2014
|
|
5YZU
 
 | |
4N1G
 
 | | Crystal Structure of Ca(2+)- discharged F88Y obelin mutant from Obelia longissima at 1.50 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-10-04 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the F88Y obelin mutant before and after bioluminescence provide molecular insight into spectral tuning among hydromedusan photoproteins
Febs J., 281, 2014
|
|
4PKX
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | Descriptor: | Protein C10C5.1, isoform i | | Authors: | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
2B07
 
 | | Crystal structure of PTP1B with Tricyclic Thiophene inhibitor. | | Descriptor: | 6-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}-3-(CARBOXYMETHOXY)THIENO[3,2-B][1]BENZOTHIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase, non-receptor type 1 | | Authors: | Moretto, A.F, Kirincich, S.J, Xu, W.X, Smith, M.J, Wan, Z.K, Wilson, D.P, Follows, B.C, Binnun, E, Joseph-McCarthy, D, Foreman, K, Erbe, D.V, Zhang, Y.L, Tam, S.K, Tam, S.Y, Lee, J. | | Deposit date: | 2005-09-13 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bicyclic and tricyclic thiophenes as protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
2AZR
 
 | | Crystal structure of PTP1B with Bicyclic Thiophene inhibitor | | Descriptor: | 3-(CARBOXYMETHOXY)THIENO[2,3-B]PYRIDINE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase, non-receptor type 1 | | Authors: | Moretto, A.F, Kirincich, S.J, Xu, W.X, Smith, M.J, Wan, Z.K, Wilson, D.P, Follows, B.C, Binnun, E, Joseph-McCarthy, D, Foreman, K, Erbe, D.V, Zhang, Y.L, Tam, S.K, Tam, S.Y, Lee, J. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bicyclic and tricyclic thiophenes as protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
4PKE
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | Descriptor: | PLATINUM (II) ION, Protein C10C5.1, isoform i | | Authors: | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | Deposit date: | 2014-05-14 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
7LKP
 
 | | Structure of ATP-free human ABCA4 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, F, Lee, J, Chen, J. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular structures of the eukaryotic retinal importer ABCA4.
Elife, 10, 2021
|
|
7LKZ
 
 | | Structure of ATP-bound human ABCA4 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Liu, F, Lee, J, Chen, J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular structures of the eukaryotic retinal importer ABCA4.
Elife, 10, 2021
|
|
4FJS
 
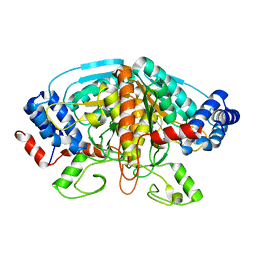 | | Crystal structure of ureidoglycolate dehydrogenase enzyme in apo form | | Descriptor: | Ureidoglycolate dehydrogenase | | Authors: | Kim, M.I, Shin, I, Lee, J, Rhee, S. | | Deposit date: | 2012-06-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
2A8C
 
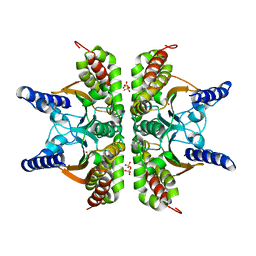 | | Haemophilus influenzae beta-carbonic anhydrase | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Lee, J, Gareiss, P.C, Preiss, J.R. | | Deposit date: | 2005-07-07 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase
Biochemistry, 45, 2006
|
|
2A8D
 
 | | Haemophilus influenzae beta-carbonic anhydrase complexed with bicarbonate | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Lee, J, Gareiss, P.C, Preiss, J.R. | | Deposit date: | 2005-07-07 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase
Biochemistry, 45, 2006
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
3E2W
 
 | |
3E2A
 
 | |
3E24
 
 | |
