1BQJ
 
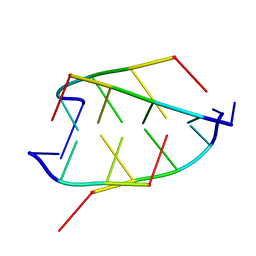 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Yang, C, Wang, S, Sutherland, C, Sinha, N, Kang, C.H. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4PPI
 
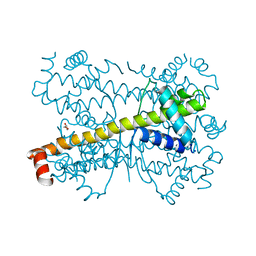 | | Crystal structure of Bcl-xL hexamer | | Descriptor: | Bcl-2-like protein 1, GLYCEROL | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural transition in Bcl-xL and its potential association with mitochondrial calcium ion transport
Sci Rep, 5, 2015
|
|
1JF4
 
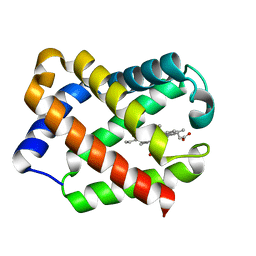 | | Crystal Structure Of Component IV Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component IV | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2001-06-20 | | Release date: | 2002-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer
Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
1JF3
 
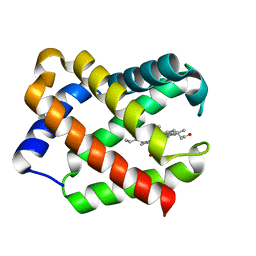 | | Crystal Structure Of Component III Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component III | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2001-06-20 | | Release date: | 2002-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer
Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AFQ
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form II) | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6L50
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 16 | | Descriptor: | 2-sulfanylidene-1,3-thiazolidin-4-one, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease.
Antiviral Res., 175, 2020
|
|
8PD6
 
 | | Crystal structure of the TRIM58 PRY-SPRY domain in complex with TRIM-473 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase TRIM58, ... | | Authors: | Renatus, M, Hoegenauer, K, Schroeder, M. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Ligands for TRIM58, a Novel Tissue-Selective E3 Ligase.
Acs Med.Chem.Lett., 14, 2023
|
|
8PD4
 
 | |
7U4B
 
 | | Ampicillin-CTX-M-15 | | Descriptor: | (2S,4S)-2-[(1S)-1-{[(2S)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase, SULFATE ION | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-02-28 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
7U57
 
 | | apo-CTX-M-15 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-03-01 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
7U48
 
 | | Clavulanic acid-CTX-M-15 | | Descriptor: | (E)-5-hydroxy-3-oxo-N-(3-oxopropylidene)-L-norvaline, Beta-lactamase, GLYCEROL, ... | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-02-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
7U49
 
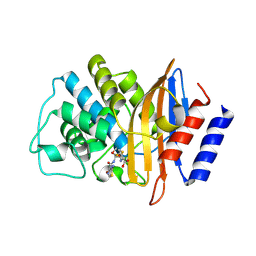 | | DFC-CTX-M-15 | | Descriptor: | (2R,4S,5R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-oxoethyl]-5-(sulfanylmethyl)-1,3-thiazinane-4-carboxylic acid, Beta-lactamase | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-02-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
5TQM
 
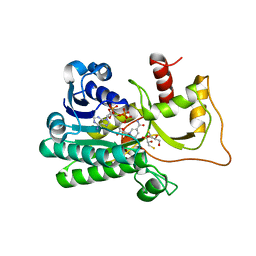 | | Cinnamoyl-CoA Reductase 1 from Sorghum bicolor in complex with NADP+ | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Cinnamoyl-CoA Reductase, GLYCEROL, ... | | Authors: | Sattler, S.A, Kang, C.H. | | Deposit date: | 2016-10-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Biochemical Characterization of Cinnamoyl-CoA Reductases.
Plant Physiol., 173, 2017
|
|
8JVE
 
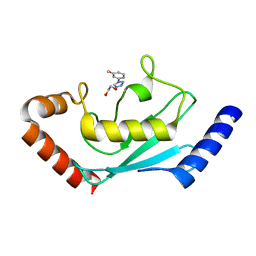 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
8JVL
 
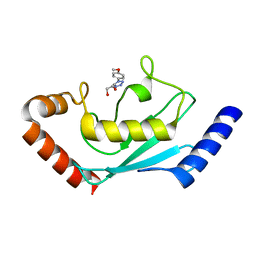 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
4EDB
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
5KHT
 
 | | Crystal structure of the N-terminal fragment of tropomyosin isoform Tpm1.1 at 1.5 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tropomyosin alpha-1 chain,General control protein GCN4 | | Authors: | Kostyukova, A.S, Krieger, I, Yoon, Y.-H, Tolkatchev, D, Samatey, F.A. | | Deposit date: | 2016-06-15 | | Release date: | 2017-06-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4964 Å) | | Cite: | Structural destabilization of tropomyosin induced by the cardiomyopathy-linked mutation R21H.
Protein Sci., 27, 2018
|
|
3HWC
 
 | |
4EDA
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
3K87
 
 | | Crystal structure of NADH:FAD oxidoreductase (TftC) - FAD complex | | Descriptor: | Chlorophenol-4-monooxygenase component 1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kang, C.H, Webb, B.N. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100.
J.Biol.Chem., 285, 2010
|
|
8IJW
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-06 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJV
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-02 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[[5-chloranyl-2-(4-chlorophenyl)phenyl]methoxy]-N-methyl-but-2-yn-1-amine, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJX
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-18 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[(5-chloranyl-2-phenylmethoxy-phenyl)methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
4W4S
 
 | | Crystal structure of ent-kaurene synthase BJKS from bradyrhizobium japonicum in complex with BPH-629 | | Descriptor: | Uncharacterized protein blr2150, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Liu, W, Zheng, Y, Huang, C.H, Guo, R.T. | | Deposit date: | 2014-08-15 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
