7DMD
 
 | |
8GVL
 
 | | PTPN21 FERM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GWH
 
 | |
8GVV
 
 | | PTPN21 PTP domain C1108S mutant | | Descriptor: | IODIDE ION, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
4B0Z
 
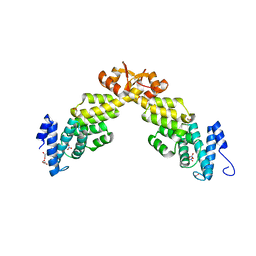 | | Crystal structure of S. pombe Rpn12 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN12, GLYCEROL, MONOTHIOGLYCEROL, ... | | Authors: | Boehringer, J, Riedinger, C, Paraskevopoulos, K, Johnson, E.O.D, Lowe, E.D, Khoudian, C, Smith, D, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-12 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural and Functional Characterisation of Rpn12 Identifies Residues Required for Rpn10 Proteasome Incorporation.
Biochem.J., 448, 2012
|
|
2KSN
 
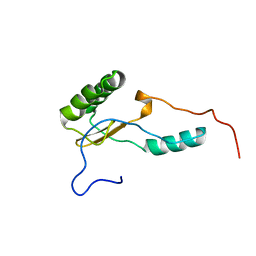 | | Solution Structure of the N-terminal Domain of DC-UbP/UBTD2 | | Descriptor: | Ubiquitin domain-containing protein 2 | | Authors: | Song, A, Zhou, C, Guan, X, Sze, K, Hu, H. | | Deposit date: | 2010-01-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of DC-UbP/UBTD2 and its interaction with ubiquitin
Protein Sci., 19, 2010
|
|
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
1UT3
 
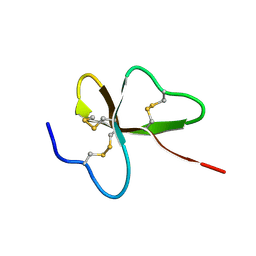 | | Solution Structure of Spheniscin-2, a beta-Defensin from Penguin Stomach Preserving Food | | Descriptor: | SPHENISCIN-2 | | Authors: | Landon, C, Labbe, H, Thouzeau, C, Bulet, P, Vovelle, F. | | Deposit date: | 2003-12-02 | | Release date: | 2004-05-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Spheniscin, a {Beta}-Defensin from the Penguin Stomach
J.Biol.Chem., 279, 2004
|
|
2VLP
 
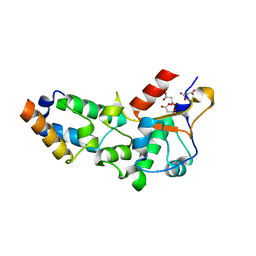 | | R54A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, MALONIC ACID | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
2VLQ
 
 | | F86A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, MALONIC ACID | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
2VLO
 
 | | K97A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, SULFATE ION | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
2VLN
 
 | | N75A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, MALONIC ACID | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
2WPT
 
 | | The crystal structure of Im2 in complex with colicin E9 DNase | | Descriptor: | COLICIN-E2 IMMUNITY PROTEIN, COLICIN-E9, GLYCEROL, ... | | Authors: | Meenan, N.A, Sharma, A, Fleishman, S.J, Macdonald, C.J, Boetzel, R, Moore, G.R, Baker, D, Kleanthous, C. | | Deposit date: | 2009-08-10 | | Release date: | 2010-06-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Structural and Energetic Basis for High Selectivity in a High-Affinity Protein-Protein Interaction.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WZ7
 
 | | Crystal structure of the N-terminal domain of E.coli YbgF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Krachler, A.M, Sharma, A, Kleanthous, C. | | Deposit date: | 2009-11-24 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Tola Modulates the Oligomeric Status of Ybgf in the Bacterial Periplasm.
J.Mol.Biol., 403, 2010
|
|
4QAY
 
 | |
3U43
 
 | |
1E0H
 
 | | Inhibitor Protein Im9 bound to its partner E9 DNase | | Descriptor: | IMMUNITY PROTEIN FOR COLICIN E9 | | Authors: | Boetzel, R, Czisch, M, Kaptein, R, Hemmings, A.M, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 2000-03-28 | | Release date: | 2000-10-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR investigation of the interaction of the inhibitor protein Im9 with its partner DNase.
Protein Sci., 9, 2000
|
|
2WQH
 
 | | Crystal structure of CTPR3Y3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Krachler, A.M, Sharma, A, Kleanthous, C. | | Deposit date: | 2009-08-21 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Self-association of TPR domains: Lessons learned from a designed, consensus-based TPR oligomer.
Proteins, 78, 2010
|
|
1E44
 
 | | ribonuclease domain of colicin E3 in complex with its immunity protein | | Descriptor: | 1,2-ETHANEDIOL, COLICIN E3, IMMUNITY PROTEIN | | Authors: | Carr, S, Walker, D, James, R, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of a Ribosome Inactivating Ribonuclease: The Crystal Structure of the Cytotoxic Domain of Colicin E3 in Complex with its Immunity Protein
Structure, 8, 2000
|
|
2XDJ
 
 | |
1EMV
 
 | | CRYSTAL STRUCTURE OF COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 (1.7 ANGSTROMS) | | Descriptor: | COLICIN E9, IMMUNITY PROTEIN IM9, PHOSPHATE ION | | Authors: | Kuhlmann, U.C, Pommer, A.J, Moore, G.M, James, R, Kleanthous, C. | | Deposit date: | 2000-03-20 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes.
J.Mol.Biol., 301, 2000
|
|
2XEV
 
 | |
1V14
 
 | |
1V15
 
 | |
1V13
 
 | |
