2IB8
 
 | |
1OU6
 
 | |
1NL7
 
 | |
1PXT
 
 | |
2IBU
 
 | |
2IBW
 
 | |
2JIG
 
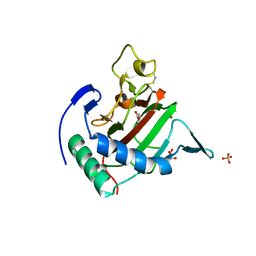 | | Crystal structure of Chlamydomonas reinhardtii prolyl-4 hydroxylase type I complexed with zinc and pyridine-2,4-dicarboxylate | | Descriptor: | GLYCEROL, PROLYL-4 HYDROXYLASE, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | Authors: | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
2JIJ
 
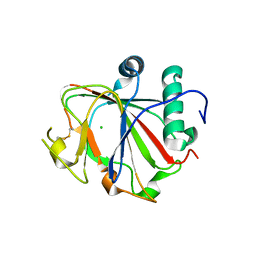 | | Crystal structure of the apo form of Chlamydomonas reinhardtii prolyl- 4 hydroxylase type I | | Descriptor: | CHLORIDE ION, PROLYL-4 HYDROXYLASE | | Authors: | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
2GCE
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (R)-IBUPROFENOYL-COENZYME A, (S)-IBUPROFENOYL-COENZYME A, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-14 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GCI
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an asparte/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (R)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-14 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GD2
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
1QFL
 
 | |
2IB9
 
 | |
2IBY
 
 | |
2IB7
 
 | |
2GD6
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
1DM3
 
 | |
1DLU
 
 | | UNLIGANDED BIOSYNTHETIC THIOLASE FROM ZOOGLOEA RAMIGERA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BIOSYNTHETIC THIOLASE, SULFATE ION | | Authors: | Modis, Y, Wierenga, R.K. | | Deposit date: | 1999-12-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase.
J.Mol.Biol., 297, 2000
|
|
1DLV
 
 | |
1E2X
 
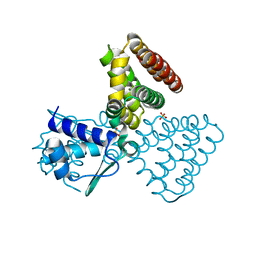 | | FadR, fatty acid responsive transcription factor from E. coli | | Descriptor: | FATTY ACID METABOLISM REGULATOR PROTEIN, SULFATE ION | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2000-05-30 | | Release date: | 2000-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Fadr, a Fatty Acid-Responsive Transcription Factor with a Novel Acyl Coenzyme A-Binding Fold
Embo J., 19, 2000
|
|
8PF8
 
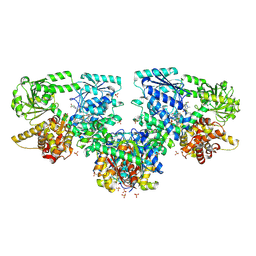 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with Fragment-M-72 | | Descriptor: | (2~{R})-3-bis[2-methyl-5-(trifluoromethyl)pyrazol-3-yl]boranyloxypropane-1,2-diol, 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystallographic fragment binding studies of the Mycobacterium tuberculosis trifunctional enzyme suggest binding pockets for the tails of the acyl-CoA substrates at its active sites and a potential substrate channeling path between them
Biorxiv, 2024
|
|
2VCY
 
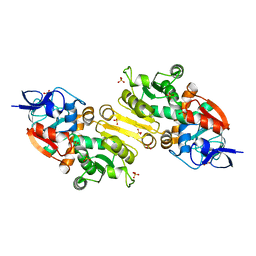 | | Crystal Structure of 2-Enoyl Thioester Reductase of Human FAS II | | Descriptor: | SULFATE ION, TRANS-2-ENOYL-COA REDUCTASE | | Authors: | Haapalainen, A.M, Pudas, R, Smart, O.S, Wierenga, R.K. | | Deposit date: | 2007-09-28 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Enzymological Studies of 2-Enoyl Thioester Reductase of the Human Mitochondrial Fas II Pathway: New Insights Into its Substrate Recognition Properties.
J.Mol.Biol., 379, 2008
|
|
2V4A
 
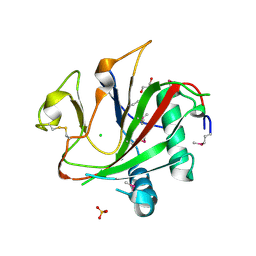 | | Crystal structure of the SeMet-labeled prolyl-4 hydroxylase (P4H) type I from green algae Chlamydomonas reinhardtii. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
2VEM
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | (3-bromo-2-oxo-propoxy)phosphonic acid, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE, TERTIARY-BUTYL ALCOHOL | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2VEN
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | CITRIC ACID, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
