1JR5
 
 | | Solution Structure of the Anti-Sigma Factor AsiA Homodimer | | Descriptor: | 10 KDA Anti-Sigma Factor | | Authors: | Urbauer, J.L, Simeonov, M.F, Bieber Urbauer, R.J, Adelman, K, Gilmore, J.M, Brody, E.N. | | Deposit date: | 2001-08-10 | | Release date: | 2002-02-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the anti-sigma factor AsiA: implications for novel functions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1ONZ
 
 | | Oxalyl-aryl-Amino Benzoic acid Inhibitors of PTP1B, compound 8b | | Descriptor: | 2-[(7-HYDROXY-NAPHTHALEN-1-YL)-OXALYL-AMINO]-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
1J6Y
 
 | | Solution structure of Pin1At from Arabidopsis thaliana | | Descriptor: | peptidyl-prolyl cis-trans isomerase | | Authors: | Landrieu, I, Wieruszeski, J.M, Wintjens, R, Inze, D, Lippens, G. | | Deposit date: | 2001-05-15 | | Release date: | 2002-08-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the
Single-domain Prolyl Cis/Trans
Isomerase PIN1At from Arabidopsis thaliana
J.Mol.Biol., 320, 2002
|
|
3FPT
 
 | | The Crystal Structure of the Complex between Evasin-1 and CCL3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Evasin-1 | | Authors: | Shaw, J.P, Dias, J.M. | | Deposit date: | 2009-01-06 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3
Plos One, 4, 2009
|
|
3FPU
 
 | |
1HI0
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus initiation complex | | Descriptor: | DNA (5'-(*TP*TP*TP*CP*C)-3'), GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-31 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
1HIL
 
 | |
1M35
 
 | | Aminopeptidase P from Escherichia coli | | Descriptor: | AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Graham, S.C, Lee, M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-06-27 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthorhombic form of Escherichia coli aminopeptidase P at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1J2O
 
 | | Structure of FLIN2, a complex containing the N-terminal LIM domain of LMO2 and ldb1-LID | | Descriptor: | Fusion of Rhombotin-2 and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
1MEY
 
 | | CRYSTAL STRUCTURE OF A DESIGNED ZINC FINGER PROTEIN BOUND TO DNA | | Descriptor: | CHLORIDE ION, CONSENSUS ZINC FINGER, DNA (5'-D(*AP*TP*GP*AP*GP*GP*CP*AP*GP*AP*AP*CP*T)-3'), ... | | Authors: | Kim, C.A, Berg, J.M. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A 2.2 A Resolution Crystal Structure of a Designed Zinc Finger Protein Bound to DNA
Nat.Struct.Biol., 3, 1996
|
|
6S1J
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(3~{S})-3-fluoranylpyrrolidin-1-yl]pyrimidin-4-yl]pyrazolo[1,5-b]pyridazine, DIMETHYL SULFOXIDE, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S11
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 6-pyridin-4-yl-3-[3-(trifluoromethyloxy)phenyl]imidazo[1,2-b]pyridazine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S14
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ~{N}-cyclopropyl-~{N}-methyl-4-pyrazolo[1,5-b]pyridazin-3-yl-pyrimidin-2-amine | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S17
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S1H
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S4C
 
 | | Crystal Structure of the vWFA2 subdomain of type VII collagen | | Descriptor: | Collagen alpha-1(VII) chain, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gebauer, J.M, Flachsenberg, F, Baumann, U, Seeger, K. | | Deposit date: | 2019-06-27 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biophysical characterization of the type VII collagen vWFA2 subdomain leads to identification of two binding sites.
Febs Open Bio, 10, 2020
|
|
6RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT WITHOUT NUCLEOTIDE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
6S8Y
 
 | | Crystal structure of cytochrome c in complex with a sulfonated quinoline-derived foldamer | | Descriptor: | 8-acetamido-2-[[2-[[2-[[2-[[2-[[2-[[2-[(2-carboxy-4-sulfonato-quinolin-8-yl)carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]quinoline-4-sulfonate, ACETATE ION, Cytochrome c iso-1, ... | | Authors: | Alex, J.M, Corvaglia, V, Hu, X, Engilberge, S, Huc, I, Crowley, P.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a protein-aromatic foldamer composite: macromolecular chiral resolution.
Chem.Commun.(Camb.), 55, 2019
|
|
6SKF
 
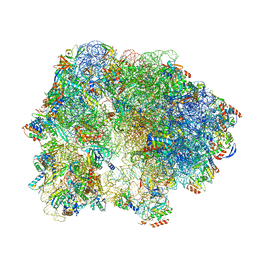 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-08-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
1BJT
 
 | | TOPOISOMERASE II RESIDUES 409-1201 | | Descriptor: | TOPOISOMERASE II | | Authors: | Fass, D, Bogden, C.E, Berger, J.M. | | Deposit date: | 1998-06-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands.
Nat.Struct.Biol., 6, 1999
|
|
6SOR
 
 | |
6SI9
 
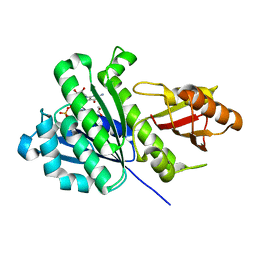 | | FtsZ-refold | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fernandez-Tornero, C, Andreu, J.M, Ruiz, F.M. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus.
Febs J., 287, 2020
|
|
1BL8
 
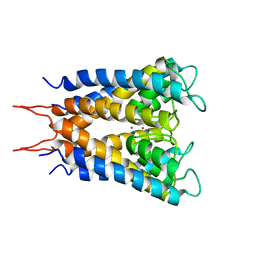 | | POTASSIUM CHANNEL (KCSA) FROM STREPTOMYCES LIVIDANS | | Descriptor: | POTASSIUM ION, PROTEIN (POTASSIUM CHANNEL PROTEIN) | | Authors: | Doyle, D.A, Cabral, J.M, Pfuetzner, R.A, Kuo, A, Gulbis, J.M, Cohen, S.L, Chait, B.T, Mackinnon, R. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the potassium channel: molecular basis of K+ conduction and selectivity.
Science, 280, 1998
|
|
6SOO
 
 | |
6SP1
 
 | | KEAP1 IN COMPLEX WITH COMPOUND 6 | | Descriptor: | (1~{S},2~{R})-2-[[(1~{S})-1-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]-5-(2-hydroxyethyloxy)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]cyclohexane-1-carboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Ontoria, J.M, Biancofiore, I, Fezzardi, P, Torrente de Haro, E, Colarusso, S, Bianchi, E, Andreini, M, Patsilinakos, A, Summa, V, Pacifici, R, Munoz-Sanjuan, I, Park, L, Bresciani, A, Dominguez, C, Toledo-Sherman, L, Harper, S. | | Deposit date: | 2019-08-30 | | Release date: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Combined Peptide and Small-Molecule Approach toward Nonacidic THIQ Inhibitors of the KEAP1/NRF2 Interaction.
Acs Med.Chem.Lett., 11, 2020
|
|
