7YRF
 
 | |
7YV7
 
 | |
7YV2
 
 | |
7WSW
 
 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
2F2O
 
 | |
3ONJ
 
 | |
7XIV
 
 | | Structural insight into the interactions between Lloviu virus VP30 and nucleoprotein | | Descriptor: | Nucleocapsid protein,Minor nucleoprotein VP30 | | Authors: | Dong, S.S, Qin, X.C, Sun, W.Y, Luan, F.C, Wang, J.J, Ma, L, Li, X.X, Yang, G.X, Hao, C.Y. | | Deposit date: | 2022-04-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structural insights into the interactions between lloviu virus VP30 and nucleoprotein.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
2GUB
 
 | |
2GLK
 
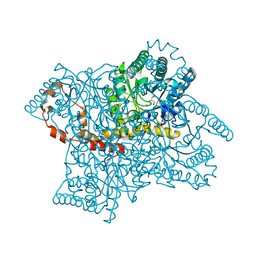 | | High-resolution study of D-Xylose isomerase, 0.94A resolution. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Carrell, H.L, Hanson, B.L, Harp, J.M, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8IS4
 
 | | Structure of an Isocytosine specific deaminase Vcz in complexed with 5-FU | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Guo, W.T, Li, X.J, Wu, B.X. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
5Y8E
 
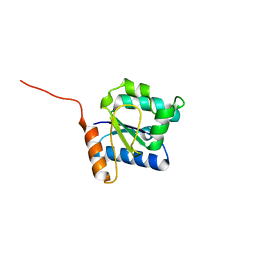 | |
5Y8F
 
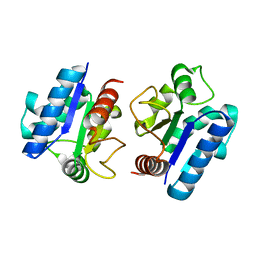 | |
3LGA
 
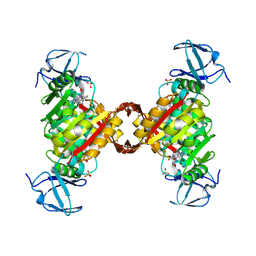 | |
5ZTK
 
 | | Synchrotron structure of light-driven chloride pump having an NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Park, S.Y, Lee, W. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency
J. Biol. Chem., 294, 2019
|
|
5ZTL
 
 | | Non-cryogenic structure of light-driven chloride pump having an NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, S.Y, Liu, H, Lee, W. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency
J. Biol. Chem., 294, 2019
|
|
4DT1
 
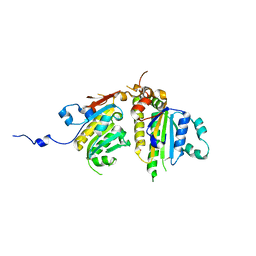 | | Crystal structure of the Psy3-Csm2 complex | | Descriptor: | Chromosome segregation in meiosis protein 2, ETHANOL, Platinum sensitivity protein 3 | | Authors: | Tao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-02-20 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural analysis of Shu proteins reveals a DNA binding role essential for resisting damage
J.Biol.Chem., 287, 2012
|
|
8JP3
 
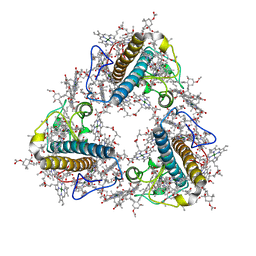 | | FCP trimer in diatom Thalassiosira pseudonana | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, ... | | Authors: | Feng, Y, Li, Z, Zhou, C.C, Liu, C, Shen, J.R, Wang, W. | | Deposit date: | 2023-06-10 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 5, 2024
|
|
5ZVS
 
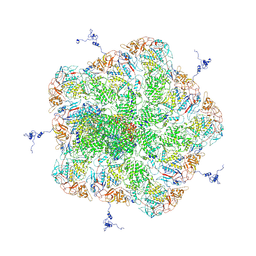 | |
9DQP
 
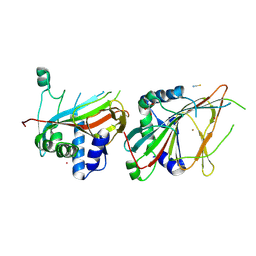 | |
9DQQ
 
 | |
9DQ1
 
 | | Crystal structure of HrmJ from Streptomyces sp. CFMR 7 (HrmJ-ssc) complexed with manganese (II), 2-oxoglutarate and 6-nitronorleucine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 6-nitro-L-norleucine, ... | | Authors: | Zheng, Y.-C, Swartz, P, Chang, W.-C. | | Deposit date: | 2024-09-23 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of a Nonheme Iron Cyclopropanase with a Homologous Hydroxylase Reveals Mechanistic Features Associated with Distinct Reaction Outcomes.
J.Am.Chem.Soc., 147, 2025
|
|
9DQ2
 
 | |
9DQR
 
 | |
9DQ0
 
 | | Crystal structure of apo HrmJ from Streptomyces sp. CFMR 7 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, BsmA domain containing protein, ... | | Authors: | Zheng, Y.-C, Chang, W.-C, Swartz, P. | | Deposit date: | 2024-09-23 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of a Nonheme Iron Cyclopropanase with a Homologous Hydroxylase Reveals Mechanistic Features Associated with Distinct Reaction Outcomes.
J.Am.Chem.Soc., 147, 2025
|
|
9DEC
 
 | | Crystal Structure of D9-threaded | | Descriptor: | D9-threaded | | Authors: | Bera, A.K, Sims, J, Baker, D. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Massively parallel assessment of designed protein solution properties using mass spectrometry and peptide barcoding.
Biorxiv, 2025
|
|
