8AAI
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with fragment E5 | | Descriptor: | (2~{S})-2-[[(2~{S})-3-methyl-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)butanoyl]amino]propanoic acid, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein-Protein Interaction: A Semi-Automated "Hit Identification-to-Optimization" Approach.
J.Med.Chem., 66, 2023
|
|
6GYH
 
 | | Crystal structure of the light-driven proton pump Coccomyxa subellipsoidea Rhodopsin CsR | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Family A G protein-coupled receptor-like protein, ... | | Authors: | Szczepek, M, Schmidt, A, Scheerer, P. | | Deposit date: | 2018-06-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a light-gated proton channel based on the crystal structure ofCoccomyxarhodopsin.
Sci.Signal., 12, 2019
|
|
3C39
 
 | | Crystal Structure of human phosphoglycerate kinase bound to 3-phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate kinase 1 | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
6DKU
 
 | | Crystal structure of Myotis VP35 mutant of interferon inhibitory domain | | Descriptor: | VP35 | | Authors: | Liu, H, Ginell, G.M, Keefe, L.J, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conservation of Structure and Immune Antagonist Functions of Filoviral VP35 Homologs Present in Microbat Genomes.
Cell Rep, 24, 2018
|
|
3C3C
 
 | | Crystal Structure of human phosphoglycerate kinase bound to 3-phosphoglycerate and L-CDP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
1BJI
 
 | |
2OAE
 
 | | Crystal structure of rat dipeptidyl peptidase (DPPIV) with thiazole-based peptide mimetic #31 | | Descriptor: | Dipeptidyl peptidase 4, N-{[(3S,5S)-5-(1,3-THIAZOLIDIN-3-YLCARBONYL)PYRROLIDIN-3-YL]METHYL}-1,3-THIAZOLE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Longenecker, K.L, Shuai, Q, Patel, J, Wiedeman, P. | | Deposit date: | 2006-12-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pyrrolidine-constrained phenethylamines: The design of potent, selective, and pharmacologically efficacious dipeptidyl peptidase IV (DPP4) inhibitors from a lead-like screening hit.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7S1W
 
 | | The AAVrh.10-glycan complex | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2021-09-02 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
3CBG
 
 | | Functional and Structural Characterization of a Cationdependent O-Methyltransferase from the Cyanobacterium Synechocystis Sp. Strain PCC 6803 | | Descriptor: | (2E)-3-(3-hydroxy-4-methoxyphenyl)prop-2-enoic acid, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, ... | | Authors: | Kopycki, J.G, Neumann, P, Stubbs, M.T. | | Deposit date: | 2008-02-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Characterization of a Cation-dependent O-Methyltransferase from the Cyanobacterium Synechocystis sp. Strain PCC 6803
J.Biol.Chem., 283, 2008
|
|
8SZ3
 
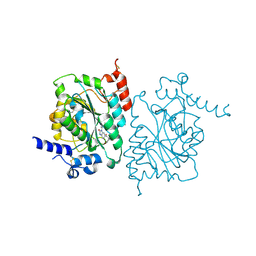 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 7j | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1S)-1-(5-bromopyridin-2-yl)ethyl]-3-[(2R)-3,3-dimethylbutan-2-yl]-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxamide, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-05-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
2QLE
 
 | | GFP/S205V mutant | | Descriptor: | Green fluorescent protein, IMIDAZOLE | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2007-07-12 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | An alternative excited-state proton transfer pathway in green fluorescent protein variant S205V.
Protein Sci., 16, 2007
|
|
2BRE
 
 | | STRUCTURE OF A HSP90 INHIBITOR BOUND TO THE N-TERMINUS OF YEAST HSP90. | | Descriptor: | 4-{4-[4-(3-AMINOPROPOXY)PHENYL]-1H-PYRAZOL-5-YL}-6-CHLOROBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
6V1Z
 
 | | genome-containing AAVrh.39 particles | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-21 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
2BRC
 
 | | Structure of a Hsp90 Inhibitor bound to the N-terminus of Yeast Hsp90. | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
6V12
 
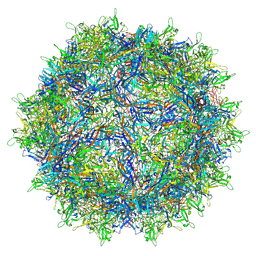 | | Empty AAV8 particles | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6V10
 
 | | genome-containing AAV8 particles | | Descriptor: | Capsid protein, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
8TU2
 
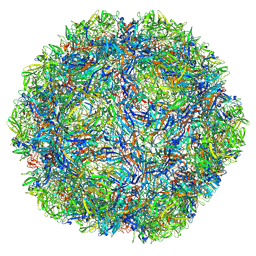 | | The Capsid of Rat Bocavirus | | Descriptor: | VP2 | | Authors: | Velez, M, Mietzsch, M, McKenna, R. | | Deposit date: | 2023-08-15 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural Characterization of Canine Minute Virus, Rat and Porcine Bocavirus.
Viruses, 15, 2023
|
|
7UMZ
 
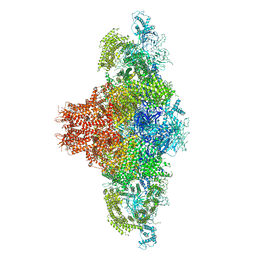 | |
8TU1
 
 | |
8TU0
 
 | |
6V1G
 
 | | Genome-containing AAVrh.10 | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
2CGF
 
 | | A RADICICOL ANALOGUE BOUND TO THE ATP BINDING SITE OF THE N-TERMINAL DOMAIN OF THE YEAST HSP90 CHAPERONE | | Descriptor: | (5Z)-13-CHLORO-14,16-DIHYDROXY-3,4,7,8,9,10-HEXAHYDRO-1H-2-BENZOXACYCLOTETRADECINE-1,11(12H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H, Moody, C.J. | | Deposit date: | 2006-03-02 | | Release date: | 2006-11-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
6V1T
 
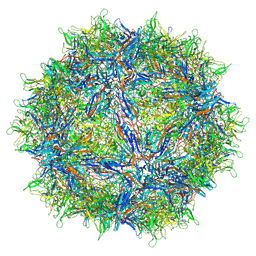 | | Empty AAVrh.39 particle | | Descriptor: | Capsid protein VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-21 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
8TIC
 
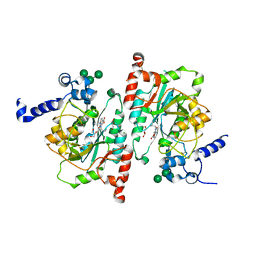 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-methylpropyl)-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxylic acid, CHLORIDE ION, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-19 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
8TJC
 
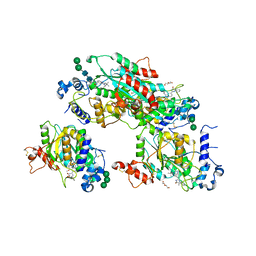 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 8a | | Descriptor: | (6M)-1-[(2R)-3,3-dimethylbutan-2-yl]-6-[(5S)-5-methyl-4-oxo-5-phenyl-4,5-dihydro-1H-imidazol-2-yl]-1,3-dihydro-2H-benzimidazol-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-20 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
