7DWC
 
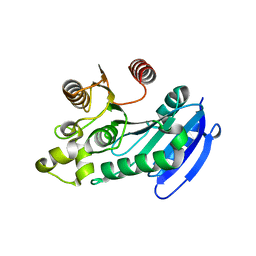 | | Bacteroides thetaiotaomicron VPI5482 BTAxe1 | | Descriptor: | Xylanase | | Authors: | Wang, L.Y, Wang, Y.L, Xin, F.J, Sun, L.C. | | Deposit date: | 2021-01-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Rational Design for Broadened Substrate Specificity and Enhanced Activity of a Novel Acetyl Xylan Esterase from Bacteroides thetaiotaomicron.
J.Agric.Food Chem., 69, 2021
|
|
1P5T
 
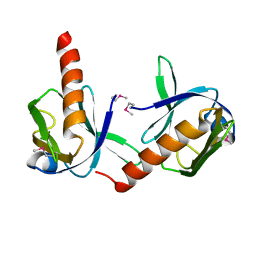 | | Crystal Structure of Dok1 PTB Domain | | Descriptor: | Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yuan, J, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.BIOL.CHEM., 279, 2004
|
|
4IZY
 
 | |
3MO0
 
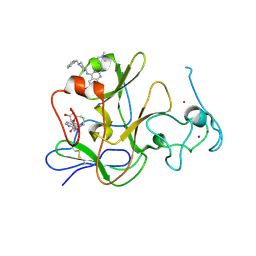 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E11 | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
6JWE
 
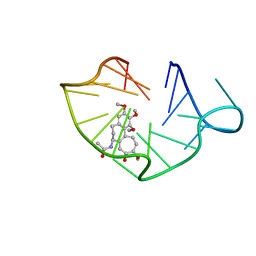 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
5DBG
 
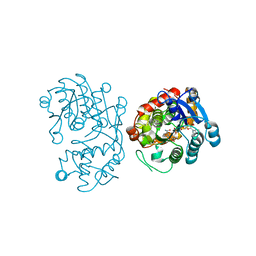 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NAD+ | | Descriptor: | Iridoid synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, W.D, Hu, Y.M, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6OGY
 
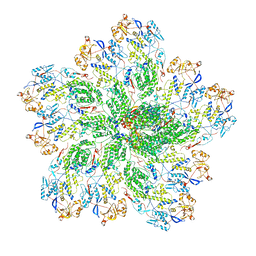 | | In situ structure of Rotavirus RNA-dependent RNA polymerase at duplex-open state | | Descriptor: | DNA/RNA (5'-D(*(GTG))-R(P*GP*C)-3'), Inner capsid protein VP2, RNA (5'-R(P*AP*GP*CP*C)-3'), ... | | Authors: | Ding, K, Chang, T, Shen, W, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release.
Nat Commun, 10, 2019
|
|
4OT6
 
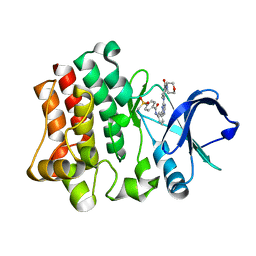 | | Crystal structure of BTK kinase domain complexed with 4-Methanesulfonyl-N-(3-{8-[4-(morpholine-4-carbonyl)-phenylamino]-imidazo[1,2-a]pyrazin-6-yl}-phenyl)-benzamide | | Descriptor: | 4-(methylsulfonyl)-N-[3-(8-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}imidazo[1,2-a]pyrazin-6-yl)phenyl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Drug Design of RN486, a Potent and Selective Bruton's Tyrosine Kinase (BTK) Inhibitor, for the Treatment of Rheumatoid Arthritis.
J.Med.Chem., 58, 2015
|
|
4OTR
 
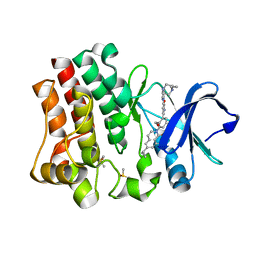 | | Crystal structure of BTK kinase domain complexed with 6-cyclopropyl-2-[3-[5-[[5-(4-ethylpiperazin-1-yl)-2-pyridyl]amino]-1-methyl-6-oxo-3-pyridyl]-2-(hydroxymethyl)phenyl]-8-fluoro-isoquinolin-1-one | | Descriptor: | 6-cyclopropyl-2-[3-(5-{[5-(4-ethylpiperazin-1-yl)pyridin-2-yl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-2-(hydroxymethyl)phenyl]-8-fluoroisoquinolin-1(2H)-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-02-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Drug Design of RN486, a Potent and Selective Bruton's Tyrosine Kinase (BTK) Inhibitor, for the Treatment of Rheumatoid Arthritis.
J.Med.Chem., 58, 2015
|
|
4OT5
 
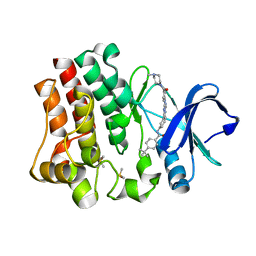 | | Crystal structure of BTK kinase domain complexed with 4-tert-Butyl-N-(3-{8-[4-(4-methyl-piperazine-1-carbonyl)-phenylamino]-imidazo[1,2-a]pyrazin-6-yl}-phenyl)-benzamide | | Descriptor: | 4-tert-butyl-N-{3-[8-({4-[(4-methylpiperazin-1-yl)carbonyl]phenyl}amino)imidazo[1,2-a]pyrazin-6-yl]phenyl}benzamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Drug Design of RN486, a Potent and Selective Bruton's Tyrosine Kinase (BTK) Inhibitor, for the Treatment of Rheumatoid Arthritis.
J.Med.Chem., 58, 2015
|
|
4OTQ
 
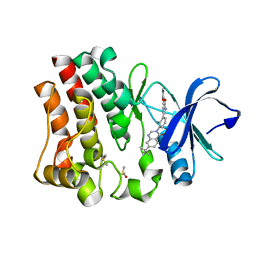 | | Crystal structure of BTK kinase domain complexed with 1-[5-[3-(7-tert-butyl-4-oxo-quinazolin-3-yl)-2-methyl-phenyl]-1-methyl-2-oxo-3-pyridyl]-3-methyl-urea | | Descriptor: | 1-{5-[3-(7-tert-butyl-4-oxoquinazolin-3(4H)-yl)-2-methylphenyl]-1-methyl-2-oxo-1,2-dihydropyridin-3-yl}-3-methylurea, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-02-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Drug Design of RN486, a Potent and Selective Bruton's Tyrosine Kinase (BTK) Inhibitor, for the Treatment of Rheumatoid Arthritis.
J.Med.Chem., 58, 2015
|
|
7XL5
 
 | | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase | | Descriptor: | NADP-dependent isopropanol dehydrogenase | | Authors: | Jiang, Y.Y, Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Engineering the hydrogen transfer pathway of an alcohol dehydrogenase to increase activity by rational enzyme design
Mol Catal, 530, 2022
|
|
1X0V
 
 | |
1X0X
 
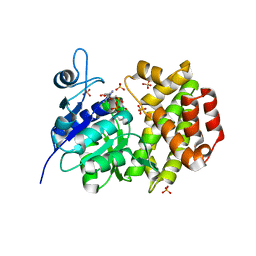 | |
7XVJ
 
 | | Crystal structure of CdpNPT in complex with harmol | | Descriptor: | 1-methyl-9~{H}-pyrido[3,4-b]indol-7-ol, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-05-24 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic formation of a prenyl beta-carboline by a fungal indole prenyltransferase.
J Nat Med, 76, 2022
|
|
7Y3V
 
 | | Crystal structure of CdpNPT in complex with harmane | | Descriptor: | 1-methyl-9H-pyrido[3,4-b]indole, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-06-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Catalytic potential of a fungal indole prenyltransferase toward beta-carbolines, harmine and harman, and their prenylation effects on antibacterial activity.
J.Biosci.Bioeng., 134, 2022
|
|
6ABS
 
 | | Actin interacting protein 5 (Aip5, mutant) | | Descriptor: | Actin binding protein, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Sun, J, Ying, X, Toh, J, Hong, W, Miao, Y, Gao, Y.G. | | Deposit date: | 2018-07-23 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization.
Nat Commun, 10, 2019
|
|
7V6F
 
 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with G3P | | Descriptor: | Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ZINC ION | | Authors: | Hongxuan, C, Huang, Y, Han, C, Chen, W, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
8D4L
 
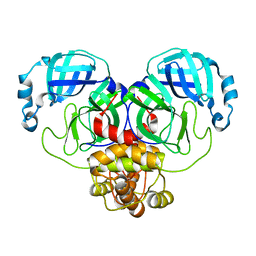 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4K
 
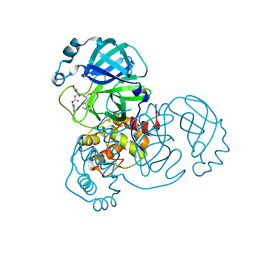 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4M
 
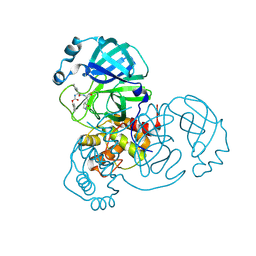 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4N
 
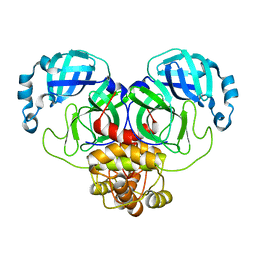 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166Q Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4J
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
6KUU
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
