8S52
 
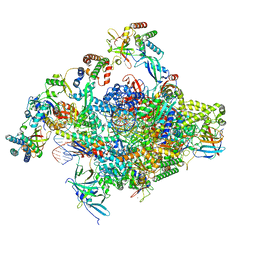 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 10 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8RM7
 
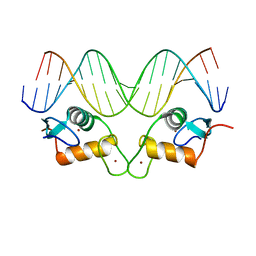 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: MMTV-177 GRE/ARE | | Descriptor: | Isoform 2 of Androgen receptor, MMTV-177 GRE/ARE Chain C, MMTV-177 GRE/ARE, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
8RXR
 
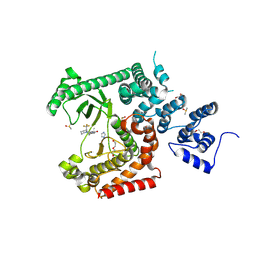 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
8RTH
 
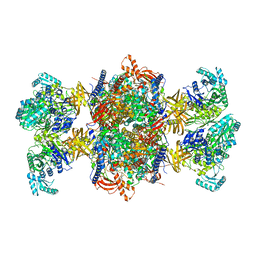 | | Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, putative, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ... | | Authors: | Ruiz, F.M, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2024-01-26 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of trypanosome 3-methylcrotonyl-CoA carboxylase provides mechanistic and dynamic insights into its enzymatic function.
Structure, 32, 2024
|
|
8RFE
 
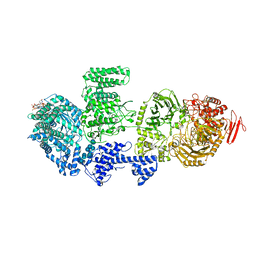 | | CgsiGP2 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase, URIDINE-5'-DIPHOSPHATE-GLUCOSE, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
8S54
 
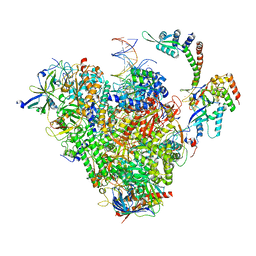 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8RFG
 
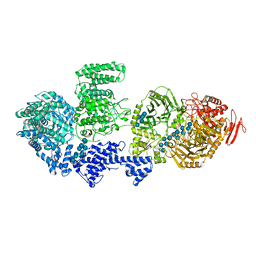 | | CgsiGP3 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, ... | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
8RF0
 
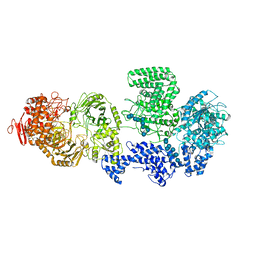 | | WT-CGS sample in nanodisc | | Descriptor: | Cyclic beta-(1,2)-glucan synthase NdvB, URIDINE-5'-DIPHOSPHATE-GLUCOSE, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, ... | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
8RBV
 
 | | SARS-CoV-2 Spike-derived peptide S976-984 S982A mutant (VLNDILARL) presented by HLA-A*02:01 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8REF
 
 | | Crystal structure of HLA B*13:01 in complex with SVLNDILARL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RH6
 
 | | Crystal structure of HLA-A*11:01 in complex with SVLNDILSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RBU
 
 | | Crystal structure of HLA-A*11:01 in complex with SVLNDILARL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RCG
 
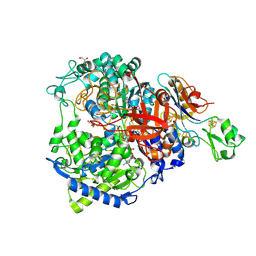 | | W-formate dehydrogenase M405S from Desulfovibrio vulgaris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural and biochemical characterization of the M405S variant of Desulfovibrio vulgaris formate dehydrogenase.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8RCV
 
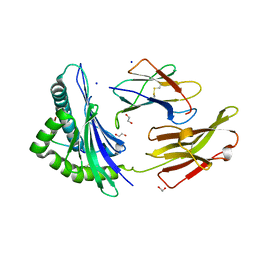 | | Crystal structure of HLA B*13:01 in complex with SVLNDIFSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen B alpha chain, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RW3
 
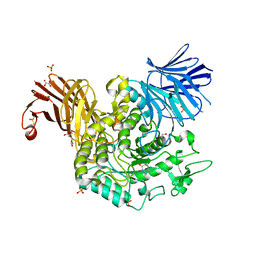 | | Crystal Structure of Agd31B, alpha-transglucosylase, complexed with a non-covalent 1,2- Cyclophellitol aziridine | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Moran, E, Davies, G, Ofamn, T, Heming, J, Nin-Hill, A, Kullmer, F, Steneker, R, Klein, A, Bennett, M, Ruijgrok, G, Kok, K, Aerts, J, Van der Marel, G, Rovira, C, Artola, M, Codee, J, Overkleeft, H. | | Deposit date: | 2024-02-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational and Electronic Variations in 1,2- and 1,5a-Cyclophellitols and their Impact on Retaining alpha-Glucosidase Inhibition.
Chemistry, 30, 2024
|
|
8RHQ
 
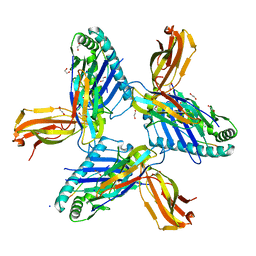 | | Crystal structure of HLA-A*11:01 in complex with SVLNDIFSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-16 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RPJ
 
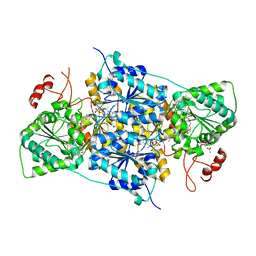 | | JanthE from Janthinobacterium sp. HH01 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RPI
 
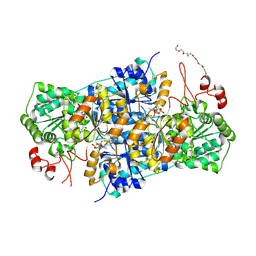 | | JanthE from Janthinobacterium sp. HH01, lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RPH
 
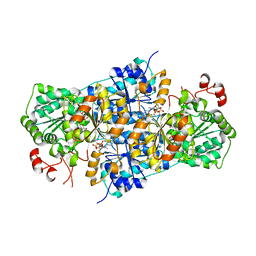 | | JanthE from Janthinobacterium sp. HH01,ketobutyryl-ThDP | | Descriptor: | (2~{S})-2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-2-yl]-2-oxidanyl-butanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6PRZ
 
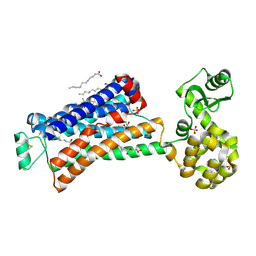 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Alprenolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
8RCB
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Reoxidized by exposure to air (in a not degassed drop) for 34 min in the presence of Formate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RCC
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - aerobic soaked with 48 bar CO2 for 1 min | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBON DIOXIDE, ... | | Authors: | Vilela-Alves, G, Carpentier, P, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RC8
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Exposed to air for 0 min | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RCA
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Reoxidized by exposure to air for 1 h in the absence of Formate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RC9
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Exposed to air for 2 h | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
