7MXO
 
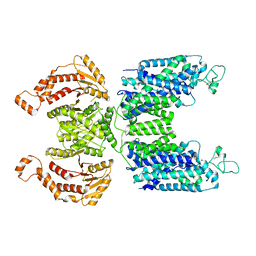 | | CryoEM structure of human NKCC1 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
7N3N
 
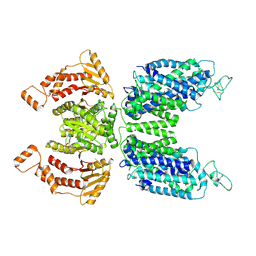 | | CryoEM structure of human NKCC1 state Fu-I | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-06-01 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
3OPO
 
 | |
3NE5
 
 | |
3OW7
 
 | |
5N8P
 
 | |
5N97
 
 | | Structure of the C. crescentus S-layer | | Descriptor: | CALCIUM ION, S-layer protein rsaA | | Authors: | Bharat, T.A, Hagen, W.J, Briggs, J.A, Lowe, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the hexagonal surface layer on Caulobacter crescentus cells.
Nat Microbiol, 2, 2017
|
|
3HGG
 
 | | Crystal Structure of CmeR Bound to Cholic Acid | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: Crystal structures of CmeR-bile acid complexes
To be Published
|
|
3K0I
 
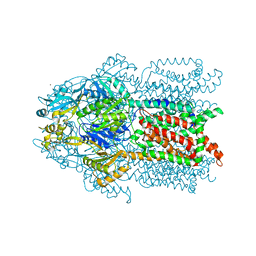 | | Crystal structure of Cu(I)CusA | | Descriptor: | COPPER (I) ION, Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.116 Å) | | Cite: | Crystal structure of CusA
To be Published
|
|
3K07
 
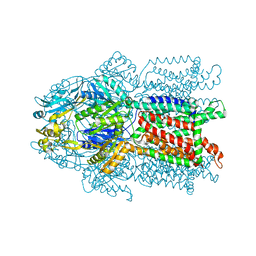 | | Crystal structure of CusA | | Descriptor: | Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.521 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
3HGY
 
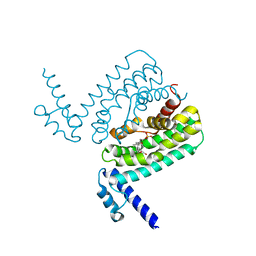 | | Crystal Structure of CmeR Bound to Taurocholic Acid | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: crystal structures of CmeR-bile acid complexes
To be Published
|
|
3KSO
 
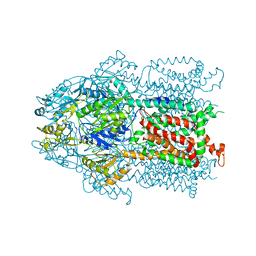 | | Structure and Mechanism of the Heavy Metal Transporter CusA | | Descriptor: | Cation efflux system protein cusA, SILVER ION | | Authors: | Su, C.-C. | | Deposit date: | 2009-11-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.367 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
7RY3
 
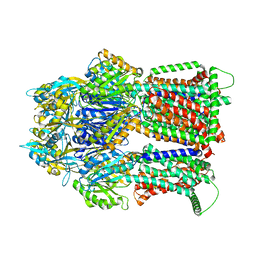 | | Multidrug Efflux pump AdeJ with TP-6076 bound | | Descriptor: | (4S,4aS,5aR,12aS)-4-(diethylamino)-3,10,12,12a-tetrahydroxy-1,11-dioxo-8-[(2S)-pyrrolidin-2-yl]-7-(trifluoromethyl)-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2021-08-24 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An Analysis of the Novel Fluorocycline TP-6076 Bound to Both the Ribosome and Multidrug Efflux Pump AdeJ from Acinetobacter baumannii.
Mbio, 13, 2022
|
|
6VQQ
 
 | |
6VKS
 
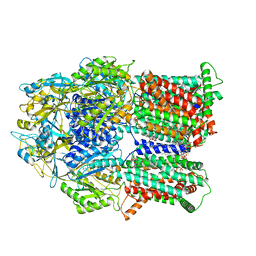 | | Cryo-electron microscopy structures of a gonococcal multidrug efflux pump illuminate a mechanism of drug recognition with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Efflux pump membrane transporter, PHOSPHATIDYLETHANOLAMINE | | Authors: | Moseng, M.A, Lyu, M. | | Deposit date: | 2020-01-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM Structures of a Gonococcal Multidrug Efflux Pump Illuminate a Mechanism of Drug Recognition and Resistance.
Mbio, 11, 2020
|
|
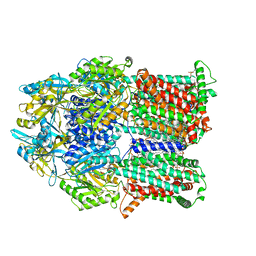
6VKT
 
 | |
6VQR
 
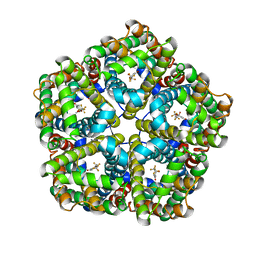 | | CryoEM Structure of the PfFNT-inhibitor complex | | Descriptor: | (2R)-2-hydroxy-7-methoxy-2-(pentafluoroethyl)-2,3-dihydro-4H-1-benzopyran-4-one, Formate-nitrite transporter | | Authors: | Su, C.C, Lyu, M. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis of transport and inhibition of the Plasmodium falciparum transporter PfFNT.
Embo Rep., 22, 2021
|
|
7M4P
 
 | | Multidrug Efflux pump AdeJ with Eravacycline bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Efflux pump membrane transporter, Eravacycline | | Authors: | Zhang, Z. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4Q
 
 | | Multidrug Efflux pump AdeJ | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
6WTI
 
 | | The Cryo-EM structure of the ubiquinol oxidase from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome o ubiquinol oxidase, ... | | Authors: | Su, C.-C. | | Deposit date: | 2020-05-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
6N40
 
 | |
6OR2
 
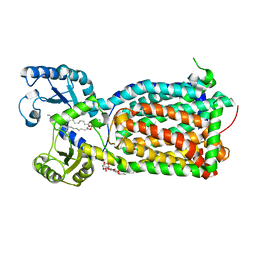 | | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, DODECYL-BETA-D-MALTOSIDE, Membrane protein, ... | | Authors: | Su, C.-C. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8SGR
 
 | | human liver mitochondrial Isovaleryl-CoA dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Isovaleryl-CoA dehydrogenase, mitochondrial | | Authors: | Zhang, Z. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8SGV
 
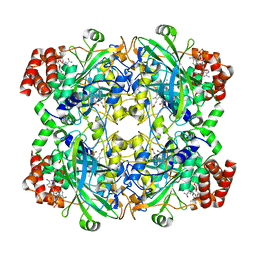 | | human liver mitochondrial Catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhang, Z. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8SGS
 
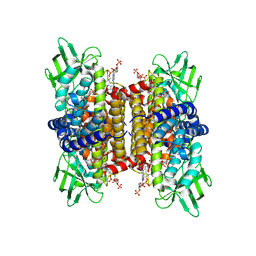 | | human liver mitochondrial Short-chain specific acyl-CoA dehydrogenase | | Descriptor: | COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, Short-chain specific acyl-CoA dehydrogenase, ... | | Authors: | Zhang, Z. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
