4QC0
 
 | | Crystal structure of human TLR8 in complex with XG-1-236 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-butyl-2H-pyrazolo[3,4-c]quinolin-4-amine, Toll-like receptor 8, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determinants of Activity at Human Toll-like Receptors 7 and 8: Quantitative Structure-Activity Relationship (QSAR) of Diverse Heterocyclic Scaffolds
J.Med.Chem., 57, 2014
|
|
4R6A
 
 | | Crystal structure of human TLR8 in complex with Hybrid-2 | | Descriptor: | 1-(4-amino-2-butyl-1H-imidazo[4,5-c]quinolin-1-yl)-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hybrid-2: AnImidazoquinoline Dual Toll-like receptor 7/8 Agonist that Potently Activates Cytokine Production by Human Newborn and Adult Leukocytes
To be Published
|
|
4QBZ
 
 | | Crystal structure of human TLR8 in complex with DS-802 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-butyl[1,3]oxazolo[4,5-c]quinolin-4-amine, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of Activity at Human Toll-like Receptors 7 and 8: Quantitative Structure-Activity Relationship (QSAR) of Diverse Heterocyclic Scaffolds
J.Med.Chem., 57, 2014
|
|
3WN4
 
 | | Crystal structure of human TLR8 in complex with DS-877 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-butylfuro[2,3-c]quinolin-4-amine, Toll-like receptor 8, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-19 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-based design of novel human Toll-like receptor 8 agonists.
Chemmedchem, 9, 2014
|
|
1ET0
 
 | | CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Nakai, T, Mizutani, H, Miyahara, I, Hirotsu, K, Takeda, S, Jhee, K.H, Yoshimura, T, Esaki, N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of 4-amino-4-deoxychorismate lyase from Escherichia coli.
J.Biochem.(Tokyo), 128, 2000
|
|
1R6X
 
 | | The Crystal Structure of a Truncated Form of Yeast ATP Sulfurylase, Lacking the C-Terminal APS Kinase-like Domain, in complex with Sulfate | | Descriptor: | ATP:sulfate adenylyltransferase, COBALT (II) ION, SULFATE ION | | Authors: | Lalor, D.J, Schnyder, T, Saridakis, V, Pilloff, D.E, Dong, A, Tang, H, Leyh, T.S, Pai, E.F. | | Deposit date: | 2003-10-17 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of a truncated form of Saccharomyces cerevisiae ATP sulfurylase: C-terminal domain essential for oligomer formation but not for activity
Protein Eng., 16, 2003
|
|
8HN3
 
 | | Soluble domain of cytochrome c-556 from Chlorobaculum tepidum | | Descriptor: | ACETATE ION, Cytochrome c-556, GLYCEROL, ... | | Authors: | Kishimoto, H, Azai, C, Yamamoto, T, Mutoh, R, Nakaniwa, T, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
8HN2
 
 | | Selenomethionine-labelled soluble domain of Rieske iron-sulfur protein from chlorobaculum tepidum | | Descriptor: | Cytochrome b6-f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Kishimoto, H, Mutoh, R, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
8FTL
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1 | | Descriptor: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-2-chloro-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]acetamide | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1
To Be Published
|
|
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFE
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD9
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8D4L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4M
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4J
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4N
 
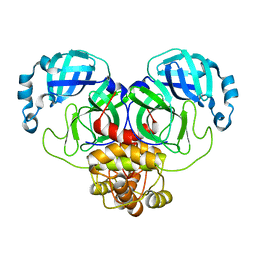 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166Q Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8EHK
 
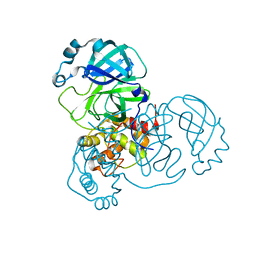 | |
8EHM
 
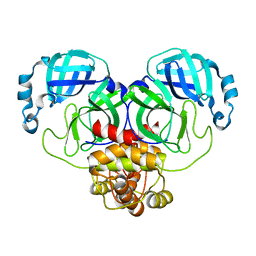 | |
8EHL
 
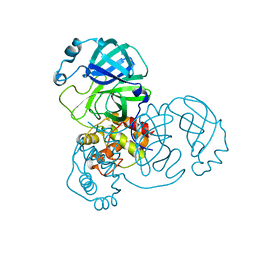 | |
8EHJ
 
 | |
7CRF
 
 | | Crystal structure of human TLR8 in complex with CU-CPD107 | | Descriptor: | 1-[2-(ethoxymethyl)-4-iodanyl-5-phenyl-imidazol-1-yl]-2-methyl-propan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Sakaniwa, K, Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2020-08-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Tetrasubstituted imidazoles as incognito Toll-like receptor 8 a(nta)gonists.
Nat Commun, 12, 2021
|
|
