7AR6
 
 | | Structure of apo SARS-CoV-2 Main Protease with large beta angle, space group C2. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AP6
 
 | | Structure of SARS-CoV-2 Main Protease bound to MUT056399. | | Descriptor: | 3C-like proteinase, 4-(4-ethyl-5-fluoranyl-2-oxidanyl-phenoxy)-3-fluoranyl-benzamide | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWS
 
 | | Structure of SARS-CoV-2 Main Protease bound to TH-302. | | Descriptor: | 3C-like proteinase, 5-[[(2-bromoethylamino)-(ethylamino)phosphoryl]oxymethyl]-1-methyl-~{N},~{N}-bis(oxidanyl)imidazol-2-amine, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AXM
 
 | | Structure of SARS-CoV-2 Main Protease bound to Pelitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6ZD9
 
 | | Crystal structure of YTHDC1 apo purified using GST tag | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD7
 
 | |
6ZD4
 
 | | Crystal structure of YTHDC1 S378A mutant | | Descriptor: | SULFATE ION, YTH domain containing 1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZCN
 
 | | Crystal structure of YTHDC1 with m6A | | Descriptor: | N6-METHYLADENOSINE-5'-MONOPHOSPHATE, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
8ZRY
 
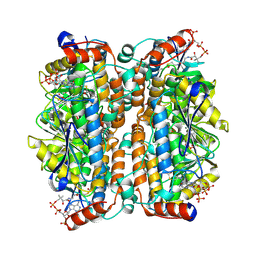 | | Structure of human ECHS1 in complex with Crotonoyl-CoA | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, MAGNESIUM ION, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRU
 
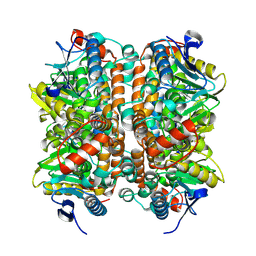 | | Structure of human ECHS1 in apo state | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, MAGNESIUM ION | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRV
 
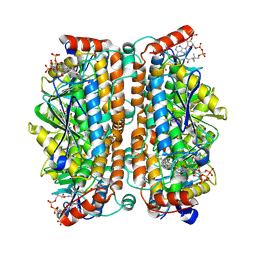 | | Structure of human ECHS1 in complex with Hexanoyl-CoA | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, HEXANOYL-COENZYME A, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRX
 
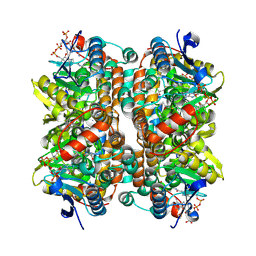 | | Structure of human ECHS1 in complex with Acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, Enoyl-CoA hydratase, mitochondrial, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRW
 
 | | Structure of human ECHS1 in complex with Octanoyl-CoA | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, MAGNESIUM ION, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8HTS
 
 | | Crystal structure of Bcl2 in complex with S-10r | | Descriptor: | 4-[2-[(2~{S})-2-(2-cyclopropylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
8HTR
 
 | | Crystal structure of Bcl2 in complex with S-9c | | Descriptor: | 4-[4-[(2~{S})-2-(2-chlorophenyl)pyrrolidin-1-yl]phenyl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
9K25
 
 | | Cryo-EM structure of apo-P2Y purinoceptor 2-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lan, B, Zhang, S, Liu, X, Lin, B. | | Deposit date: | 2024-10-17 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural insight into the self-activation and G-protein coupling of P2Y2 receptor.
Cell Discov, 11, 2025
|
|
9K0K
 
 | | Cryo-EM structure of UTP-bound P2Y purinoceptor 4-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lan, B, Zhang, S, Liu, X, Lin, B. | | Deposit date: | 2024-10-15 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insight into the self-activation and G-protein coupling of P2Y2 receptor.
Cell Discov, 11, 2025
|
|
9K0X
 
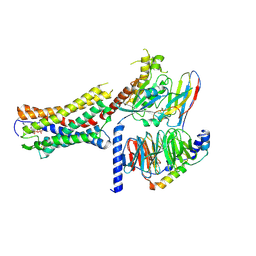 | | Cryo-EM structure of ATP-bound P2Y purinoceptor 2-miniGq-Nb35 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lan, B, Zhang, S, Liu, X, Lin, B. | | Deposit date: | 2024-10-16 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insight into the self-activation and G-protein coupling of P2Y2 receptor.
Cell Discov, 11, 2025
|
|
9K20
 
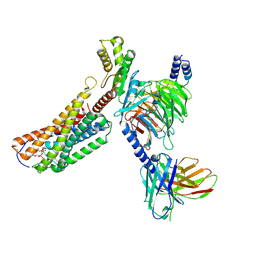 | | Cryo-EM structure of ATP-bound P2Y purinoceptor 2-miniGo-scFv16 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lan, B, Zhang, S, Liu, X, Lin, B. | | Deposit date: | 2024-10-16 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural insight into the self-activation and G-protein coupling of P2Y2 receptor.
Cell Discov, 11, 2025
|
|
1B0L
 
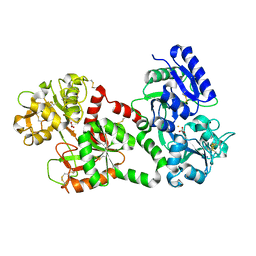 | | RECOMBINANT HUMAN DIFERRIC LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, PROTEIN (LACTOFERRIN) | | Authors: | Baker, E.N, Jameson, G.B, Sun, X. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of recombinant human lactoferrin expressed in Aspergillus awamori.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5GJT
 
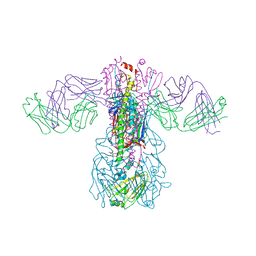 | | Crystal structure of H1 hemagglutinin from A/Washington/05/2011 in complex with a neutralizing antibody 3E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, heavy chain of human neutralizing antibody 3E1, ... | | Authors: | Wang, W, Zhang, T, Ding, J. | | Deposit date: | 2016-07-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza A viruses
Nat Commun, 7, 2016
|
|
4C13
 
 | | x-ray crystal structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ruane, K.M, Roper, D.I, Fulop, V, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy.
Nat.Chem.Biol., 2021
|
|
5GJS
 
 | | Crystal structure of H1 hemagglutinin from A/California/04/2009 in complex with a neutralizing antibody 3E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, W, Zhang, T, Ding, J. | | Deposit date: | 2016-07-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza A viruses
Nat Commun, 7, 2016
|
|
