4TQP
 
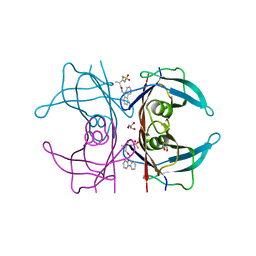 | | Human transthyretin (TTR) complexed with (R)-3-(9H-fluoren-9-ylideneaminooxy)-2-methyl-N-(methylsulfonyl) propionamide in a dual binding mode | | Descriptor: | (2R)-3-[(9H-fluoren-9-ylideneamino)oxy]-2-methyl-N-(methylsulfonyl)propanamide, GLYCEROL, Transthyretin | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
4TQI
 
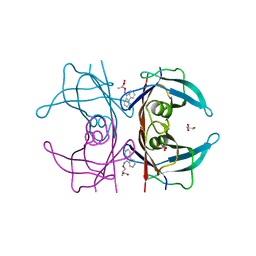 | | Human transthyretin (TTR) complexed with 3-(9H-fluoren-9-ylideneaminooxy)propanoic acid in a dual binding mode | | Descriptor: | (2S)-3-[(9H-fluoren-9-ylideneamino)oxy]-2-methylpropanoic acid, GLYCEROL, Transthyretin | | Authors: | Stura, E.A, Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
4TQ8
 
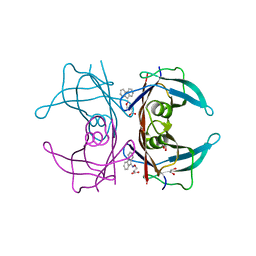 | | Dual binding mode for 3-(9H-fluoren-9-ylideneaminooxy)propanoic acid binding to Human transthyretin (TTR) | | Descriptor: | 1,2-ETHANEDIOL, 3-[(9H-fluoren-9-ylideneamino)oxy]propanoic acid, Transthyretin | | Authors: | Ciccone, L, Orlandini, E, Nencetti, S, Rossello, A, Stura, E.A. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
1GNC
 
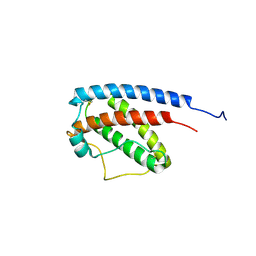 | |
3ZIZ
 
 | | Crystal structure of Podospora anserina GH5 beta-(1,4)-mannanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GH5 ENDO-BETA-1,4-MANNANASE, GLYCEROL | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-01-15 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
5T7L
 
 | | Pt(II)-mediated copper-dependent interactions between ATOX1 and MNK1 | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, Copper-transporting ATPase 1, ... | | Authors: | Caliandro, R, Mirabelli, V, Caliandro, R, Rosato, A, Lasorsa, A, Galliani, A, Arnesano, F, Natile, G. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic and Structural Basis for Inhibition of Copper Trafficking by Platinum Anticancer Drugs.
J.Am.Chem.Soc., 141, 2019
|
|
3JC1
 
 | | Electron cryo-microscopy of the IST1-CHMP1B ESCRT-III copolymer | | Descriptor: | Charged multivesicular body protein 1b, Increased Sodium Tolerance 1 (IST1) | | Authors: | McCullough, J, Clippinger, A.K, Talledge, N, Skowyra, M.L, Saunders, M.G, Naismith, T.V, Colf, L.A, Afonine, P, Arthur, C, Sundquist, W.I, Hanson, P.I, Frost, A. | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and membrane remodeling activity of ESCRT-III helical polymers.
Science, 350, 2015
|
|
4TQH
 
 | | Human transthyretin (TTR) complexed with 3-(9H-fluoren-9-ylideneaminooxy)ethanoic acid | | Descriptor: | 1,2-ETHANEDIOL, Transthyretin, [(9H-fluoren-9-ylideneamino)oxy]acetic acid | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
1W6H
 
 | | Novel plasmepsin II-inhibitor complex | | Descriptor: | N-((3S,4S)-5-[(4-BROMOBENZYL)OXY]-3-HYDROXY-4-{[N-(PYRIDIN-2-YLCARBONYL)-L-VALYL]AMINO}PENTANOYL)-L-ALANYL-L-LEUCINAMIDE, PLASMEPSIN 2 | | Authors: | Lindberg, J, Johansson, P.-O, Rosenquist, A, Kvarnstroem, I, Vrang, L, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-18 | | Release date: | 2006-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Study of a Novel Inhibitor with Bulky P1 Side Chain in Complex with Plasmepsin II -Implications for Drug Design
To be Published
|
|
1K3H
 
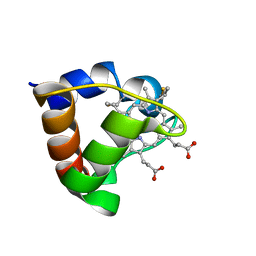 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3G
 
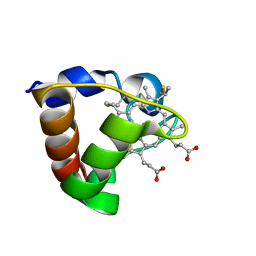 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1Y3K
 
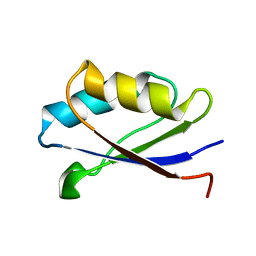 | | Solution structure of the apo form of the fifth domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Chasapis, C.T, Ciofi-Baffoni, S, Hadjiliadis, N, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-11-25 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal-binding domains of the Menkes protein
Febs J., 272, 2005
|
|
1Y3J
 
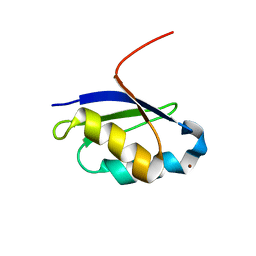 | | Solution structure of the copper(I) form of the fifth domain of Menkes protein | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Chasapis, C.T, Ciofi-Baffoni, S, Hadjiliadis, N, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-11-25 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal-binding domains of the Menkes protein
Febs J., 272, 2005
|
|
1OLL
 
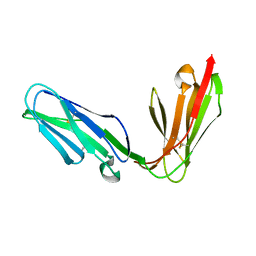 | | Extracellular region of the human receptor NKp46 | | Descriptor: | 1,2-ETHANEDIOL, NK RECEPTOR | | Authors: | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Pesce, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-08-07 | | Release date: | 2003-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the Human Nk Cell Triggering Receptor Nkp46 Ectodomain
Biochem.Biophys.Res.Commun., 309, 2003
|
|
1YJU
 
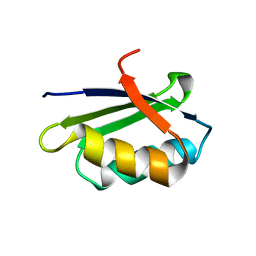 | | Solution structure of the apo form of the sixth soluble domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJT
 
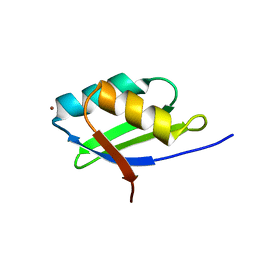 | | Solution structure of the Cu(I) form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S. | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJR
 
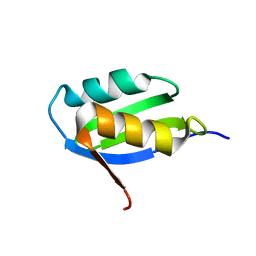 | | Solution structure of the apo form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJV
 
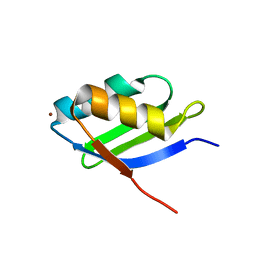 | | Solution structure of the Cu(I) form of the sixth soluble domain of Menkes protein | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S. | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
5E4A
 
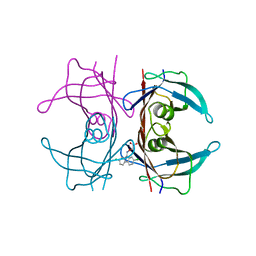 | | Human transthyretin (TTR) complexed with (2,7-Dichloro-fluoren-9-ylideneaminooxy)-acetic acid. | | Descriptor: | Transthyretin, {[(2,7-dichloro-9H-fluoren-9-ylidene)amino]oxy}acetic acid | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Synthesis and structural analysis of halogen substituted fibril formation inhibitors of Human Transthyretin (TTR).
J Enzyme Inhib Med Chem, 31, 2016
|
|
1H4K
 
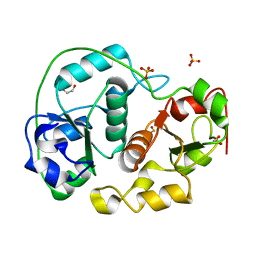 | | Sulfurtransferase from Azotobacter vinelandii in complex with hypophosphite | | Descriptor: | 1,2-ETHANEDIOL, HYPOPHOSPHITE, SULFATE ION, ... | | Authors: | Bordo, D, Forlani, F, Spallarossa, A, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Persulfurated Cysteine Promotes Active Site Reactivity in Azotobacter Vinelandii Rhodanse
Biol.Chem., 382, 2001
|
|
5E4O
 
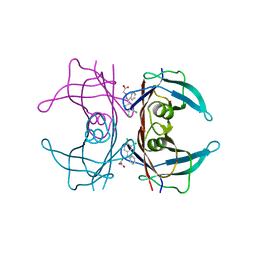 | | Human transthyretin (TTR) complexed with (Z)-((3,4-Dichloro-phenyl)-methyleneaminooxy)-acetic acid | | Descriptor: | ({(Z)-[(3,4-dichlorophenyl)(phenyl)methylidene]amino}oxy)acetic acid, Transthyretin | | Authors: | Ciccone, L, Savko, M, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2015-10-06 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and structural analysis of halogen substituted fibril formation inhibitors of Human Transthyretin (TTR).
J Enzyme Inhib Med Chem, 31, 2016
|
|
5E23
 
 | | Human transthyretin (TTR) complexed with (2,7-Dibromo-fluoren-9-ylideneaminooxy)-acetic acid | | Descriptor: | DIMETHYL SULFOXIDE, Transthyretin, {[(2,7-dibromo-9H-fluoren-9-ylidene)amino]oxy}acetic acid | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Synthesis and structural analysis of halogen substituted fibril formation inhibitors of Human Transthyretin (TTR).
J Enzyme Inhib Med Chem, 31, 2016
|
|
1HKF
 
 | | The three dimensional structure of NK cell receptor Nkp44, a triggering partner in natural cytotoxicity | | Descriptor: | NK CELL ACTIVATING RECEPTOR | | Authors: | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-03-10 | | Release date: | 2003-06-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Three-Dimensional Structure of the Human Nk Cell Receptor Nkp44, a Triggering Partner in Natural Cytotoxicity
Structure, 11, 2003
|
|
1HKS
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
1HKT
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
