9QK4
 
 | | X-ray crystal structure of SlPYL1-Ferulic Acid complex | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, DIMETHYL SULFOXIDE, SlPYL1-NIO | | Authors: | Rivera-Moreno, M, Merino-Gracia, J, Infantes, L, Albert, A. | | Deposit date: | 2025-03-19 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural modulators of abscisic acid Signaling: Insights into polyphenol-based antagonists and their role in ABA receptor regulation.
Plant Physiol Biochem., 227, 2025
|
|
9QK6
 
 | | X-ray crystal structure of SlPYL1-FeruloylPhenylalanine complex | | Descriptor: | (2~{S})-2-[[(~{E})-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enoyl]amino]-3-phenyl-propanoic acid, DIMETHYL SULFOXIDE, SlPYL1-NIO | | Authors: | Rivera-Moreno, M, Merino-Gracia, J, Infantes, L, Albert, A. | | Deposit date: | 2025-03-19 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Natural modulators of abscisic acid Signaling: Insights into polyphenol-based antagonists and their role in ABA receptor regulation.
Plant Physiol Biochem., 227, 2025
|
|
9QK5
 
 | | X-ray crystal structure of SlPYL1-Caffeic Acid complex | | Descriptor: | CAFFEIC ACID, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Rivera-Moreno, M, Merino-Gracia, J, Infantes, L, Albert, A. | | Deposit date: | 2025-03-19 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Natural modulators of abscisic acid Signaling: Insights into polyphenol-based antagonists and their role in ABA receptor regulation.
Plant Physiol Biochem., 227, 2025
|
|
3KBX
 
 | |
4RA8
 
 | | Structure analysis of the Mip1a P8A mutant | | Descriptor: | C-C motif chemokine 3 | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RAL
 
 | | Crystal structure of insulin degrading enzyme in complex with macrophage inflammatory protein 1 beta | | Descriptor: | C-C motif chemokine 4, Insulin-degrading enzyme, ZINC ION | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
2V3R
 
 | | Hypocrea jecorina Cel7A in complex with (S)-dihydroxy-phenanthrenolol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(2S)-2-HYDROXY-3-(9-PHENANTHRYLOXY)PROPYL]AMINO}PROPANE-1,3-DIOL, COBALT (II) ION, ... | | Authors: | Fagerstrom, A, Sandgren, M, Berg, U, Stahlberg, J. | | Deposit date: | 2007-06-21 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Study of the Chiral Recognition Mechanisms of Cellobiohydrolase Cel7A for Ligands Based on the Beta-Blocker Motif: Crystal Structures, Microcalorimetry and Computational Modelling of Cel7A-Inhibitor Complexes.
To be Published
|
|
2V3I
 
 | | Hypocrea jecorina Cel7A in complex with (R)-dihydroxy-phenanthrenolol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(2R)-2-HYDROXY-3-(9-PHENANTHRYLOXY)PROPYL]AMINO}PROPANE-1,3-DIOL, COBALT (II) ION, ... | | Authors: | Fagerstrom, A, Sandgren, M, Berg, U, Stahlberg, J. | | Deposit date: | 2007-06-18 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A Study of the Chiral Recognition Mechanisms of Cellobiohydrolase Cel7A for Ligands Based on the Beta-Blocker Motif: Crystal Structures, Microcalorimetry and Computational Modelling of Cel7A-Inhibitor Complexes.
To be Published
|
|
6GWS
 
 | | Crystal structure of human PCNA in complex with three p15 peptides | | Descriptor: | PCNA-associated factor, Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Gonzalez-Magana, A, Romano-Moreno, M, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
6OST
 
 | | RF2 pre-accommodated state bound Release complex 70S at 24ms | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSQ
 
 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-26 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSK
 
 | | RF1 accommodated 70S complex at 60 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-06-26 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
4NUL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: D58P OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
4NLL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57D OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
6OUO
 
 | | RF2 accommodated state bound 70S complex at long incubation time | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OT3
 
 | | RF2 accommodated state bound Release complex 70S at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
7UYU
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYR
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 12 | | Descriptor: | 2-(4-{[2-(2,6-difluorophenyl)-5-oxo-5H-pyrrolo[3,4-d]pyrimidin-4-yl]amino}phenyl)-N-ethylacetamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYW
 
 | | Crystal structure of JAK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Tyrosine-protein kinase JAK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYT
 
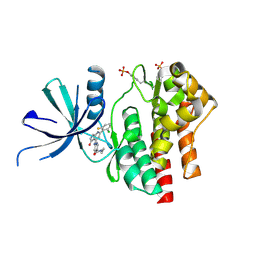 | | Crystal structure of TYK2 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYV
 
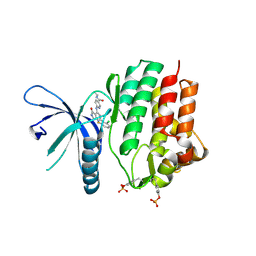 | | Crystal structure of JAK3 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, CHLORIDE ION, Tyrosine-protein kinase JAK3 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYS
 
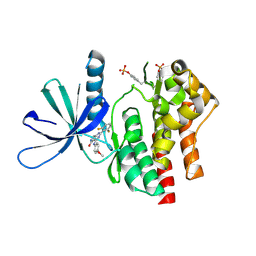 | | Crystal structure of TYK2 kinase domain in complex with compound 16 | | Descriptor: | 2-(2,6-difluorophenyl)-4-(4-methoxyanilino)-5H-pyrrolo[3,4-d]pyrimidin-5-one, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7URM
 
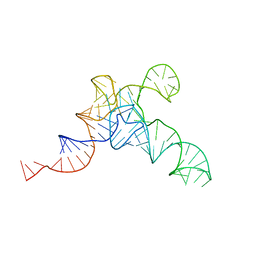 | | allo-tRNAUTu1A in the P site of the E. coli ribosome | | Descriptor: | allo-tRNAUTU1A | | Authors: | Zhang, J, Prabhakar, A, Krahn, N, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
7UR5
 
 | | allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome | | Descriptor: | allo-tRNAUTu1 | | Authors: | Zhang, J, Krahn, N, Prabhakar, A, Vargas-Rodriguez, O, Krupkin, M, Fu, Z, Acosta-Reyes, F.J, Ge, X, Choi, J, Crnkovic, A, Ehrenberg, M, Viani Puglisi, E, Soll, D, Puglisi, J. | | Deposit date: | 2022-04-21 | | Release date: | 2022-08-10 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uncovering translation roadblocks during the development of a synthetic tRNA.
Nucleic Acids Res., 50, 2022
|
|
