7MKZ
 
 | | PTP1B F225Y mutant, open state | | Descriptor: | CHLORIDE ION, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MOV
 
 | | PTP1B 1-301 F225Y-R199N mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
5JPF
 
 | | Serine/Threonine phosphatase Z1 (Candida albicans) binds to inhibitor microcystin-LR | | Descriptor: | MALONATE ION, MANGANESE (II) ION, Microcystin-LR, ... | | Authors: | Choy, M.S, Chen, E.H, Peti, W, Page, R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39929724 Å) | | Cite: | Molecular Insights into the Fungus-Specific Serine/Threonine Protein Phosphatase Z1 in Candida albicans.
Mbio, 7, 2016
|
|
5KA8
 
 | | Protein Tyrosine Phosphatase 1B L192A mutant, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
5KA1
 
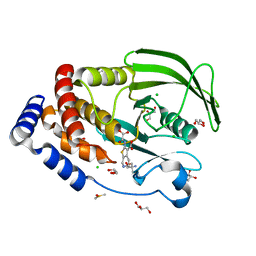 | | Protein Tyrosine Phosphatase 1B Delta helix 7 mutant in complex with TCS401, closed state | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
7MN7
 
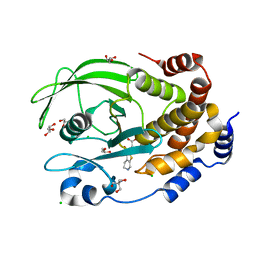 | | PTP1B F225Y in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MN9
 
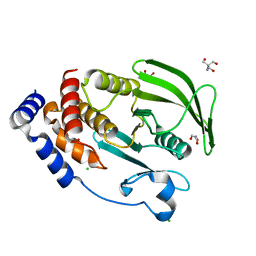 | | PTP1B 1-284 F225Y-R199N | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNA
 
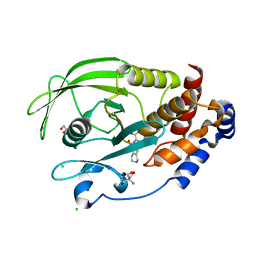 | | PTP1B 1-284 F225Y-R199N in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNC
 
 | | PTP1B L204A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MND
 
 | | PTP1B L204A in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNF
 
 | | PTP1B P206G in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNE
 
 | | PTP1B P206G mutation, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
6ZQS
 
 | | Crystal structure of double-phosphorylated p38alpha with ATF2(83-102) | | Descriptor: | 2-[(2,4-difluorophenyl)amino]-7-{[(2R)-2,3-dihydroxypropyl]oxy}-10,11-dihydro-5H-dibenzo[a,d][7]annulen-5-one, Cyclic AMP-dependent transcription factor ATF-2, Mitogen-activated protein kinase 14 | | Authors: | Kirsch, K, Sok, P, Poti, A.L, Remenyi, A. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-regulation of the transcription controlling ATF2 phosphoswitch by JNK and p38.
Nat Commun, 11, 2020
|
|
6ZR5
 
 | | Crystal structure of JNK1 in complex with ATF2(19-58) | | Descriptor: | Cyclic AMP-dependent transcription factor ATF-2, MAGNESIUM ION, Mitogen-activated protein kinase 8, ... | | Authors: | Kirsch, K, Zeke, A, Remenyi, A. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Co-regulation of the transcription controlling ATF2 phosphoswitch by JNK and p38.
Nat Commun, 11, 2020
|
|
6GHM
 
 | | Structure of PP1 alpha phosphatase bound to ASPP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Apoptosis-stimulating of p53 protein 2, ... | | Authors: | Mouilleron, S, Bertran, T.M, Tapon, N, Zhou, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ASPP proteins discriminate between PP1 catalytic subunits through their SH3 domain and the PP1 C-tail.
Nat Commun, 10, 2019
|
|
6TCA
 
 | | Phosphorylated p38 and MAPKAPK2 complex with inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14, N-[5-(dimethylsulfamoyl)-2-methylphenyl]-1-phenyl-5-propyl-1H-pyrazole-4-carboxamide | | Authors: | Sok, P, Remenyi, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | MAP Kinase-Mediated Activation of RSK1 and MK2 Substrate Kinases.
Structure, 28, 2020
|
|
2QDM
 
 | |
2QDC
 
 | |
2QDP
 
 | |
2HVL
 
 | |
7F5N
 
 | | Crystal structure of TCPTP catalytic domain | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Singh, J.P, Lin, M.-J, Hsu, S.-F, Lee, C.-C, Meng, T.-C. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of TCPTP Unravels an Allosteric Regulatory Role of Helix alpha 7 in Phosphatase Activity.
Biochemistry, 60, 2021
|
|
7F5O
 
 | | Crystal structure of PTPN2 catalytic domain | | Descriptor: | IODIDE ION, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Singh, J.P, Lin, M.-J, Hsu, S.-F, Lee, C.-C, Meng, T.-C. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of TCPTP Unravels an Allosteric Regulatory Role of Helix alpha 7 in Phosphatase Activity.
Biochemistry, 60, 2021
|
|
3D42
 
 | | Crystal structure of HePTP in complex with a monophosphorylated Erk2 peptide | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Mitogen-activated protein kinase 1 peptide, ... | | Authors: | Critton, D.A, Tortajada, A, Page, R. | | Deposit date: | 2008-05-13 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis of substrate recognition by hematopoietic tyrosine phosphatase.
Biochemistry, 47, 2008
|
|
3D44
 
 | | Crystal structure of HePTP in complex with a dually phosphorylated Erk2 peptide mimetic | | Descriptor: | CHLORIDE ION, GLYCEROL, Mitogen-activated protein kinase 1 peptide, ... | | Authors: | Critton, D.A, Tortajada, A, Page, R. | | Deposit date: | 2008-05-13 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of substrate recognition by hematopoietic tyrosine phosphatase.
Biochemistry, 47, 2008
|
|
