8TGA
 
 | |
8TG9
 
 | |
7NT4
 
 | | X-ray structure of SCoV2-PLpro in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, PROFLAVIN, ... | | Authors: | Napolitano, V, Mourao, A, Bostock, M, Matsuda, A, Czarna, A, Popowicz, G.M. | | Deposit date: | 2021-03-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Acriflavine, a clinically approved drug, inhibits SARS-CoV-2 and other betacoronaviruses.
Cell Chem Biol, 29, 2022
|
|
7NS6
 
 | | SARS-CoV-2 Spike (dimers) in complex with six Fu2 nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fu2 nanobody, Spike glycoprotein,Fibritin, ... | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A bispecific monomeric nanobody induces spike trimer dimers and neutralizes SARS-CoV-2 in vivo.
Nat Commun, 13, 2022
|
|
3CG8
 
 | | Laccase from Streptomyces coelicolor | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, TETRAETHYLENE GLYCOL, ... | | Authors: | Skalova, T, Dohnalek, J, Ostergaard, L.H, Ostergaard, P.R, Kolenko, P, Duskova, J, Hasek, J. | | Deposit date: | 2008-03-05 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | The Structure of the Small Laccase from Streptomyces coelicolor Reveals a Link between Laccases and Nitrite Reductases.
J.Mol.Biol., 385, 2009
|
|
6QCG
 
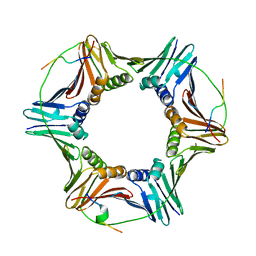 | | PCNA complex with Cdt1 N-terminal PIP-box peptide | | Descriptor: | DNA replication factor Cdt1, Proliferating cell nuclear antigen | | Authors: | Perrakis, A, von Castelmur, E. | | Deposit date: | 2018-12-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Direct binding of Cdt2 to PCNA is important for targeting the CRL4Cdt2E3 ligase activity to Cdt1.
Life Sci Alliance, 1, 2018
|
|
6QC0
 
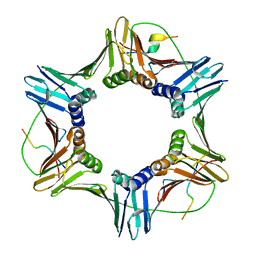 | | PCNA complex with Cdt2 C-terminal PIP-box peptide | | Descriptor: | Denticleless protein homolog, Proliferating cell nuclear antigen | | Authors: | Perrakis, A.P, von Castelmur, E. | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Direct binding of Cdt2 to PCNA is important for targeting the CRL4Cdt2E3 ligase activity to Cdt1.
Life Sci Alliance, 1, 2018
|
|
6NCO
 
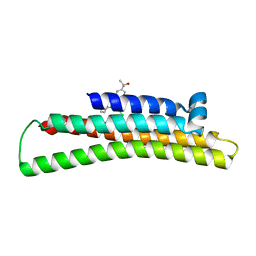 | | Fragment-based Discovery of an apoE4 Stabilizer | | Descriptor: | 1-[5-chloro-4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]cyclobutane-1-carboximidamide, Apolipoprotein E | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer.
J.Med.Chem., 62, 2019
|
|
4UOW
 
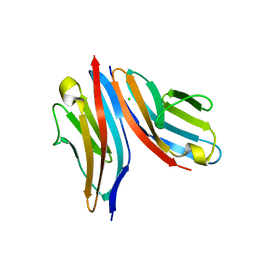 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | CHLORIDE ION, Obscurin, SODIUM ION, ... | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
6Y1K
 
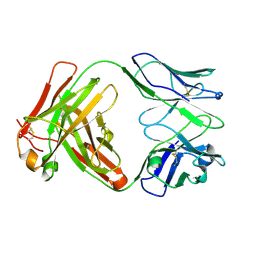 | | Crystal structure of the unmodified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | FAB A.17 L47R mutant Heavy Chain, FAB A.17 L47R mutant Light Chain | | Authors: | Chatziefthimiou, S, Stepanova, A, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8ULD
 
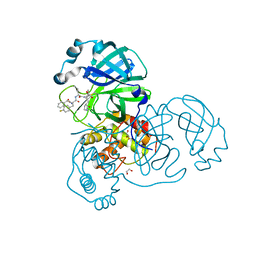 | | SARA CoV-2 3C-like protease in complex with GSK3487016A | | Descriptor: | 1,2-ETHANEDIOL, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-{(2S,3R)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]-4-[(propan-2-yl)amino]butan-2-yl}-L-leucinamide, Replicase polyprotein 1a | | Authors: | Williams, S.P, Concha, N.O. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
8UIF
 
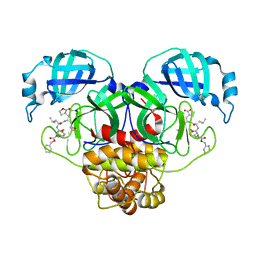 | | Crystal structure of SARS CoV-2 3CL protease in complex with GSK4365096A | | Descriptor: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-{(2R)-1-[(2S)-oxolan-2-yl]-3-[(3S)-2-oxooxolan-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Concha, N.O, Williams, S.P. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
8UHO
 
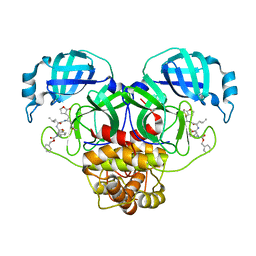 | | Crystal structure of SARS CoV-2 3CL protease in complex with GSK4365096A | | Descriptor: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-[(2S,3Z)-1-[(2S)-oxolan-2-yl]-3-(2-oxooxolan-3-ylidene)propan-2-yl]-L-leucinamide | | Authors: | Concha, N.O, Williams, S.P. | | Deposit date: | 2023-10-09 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
8UIA
 
 | |
8RKI
 
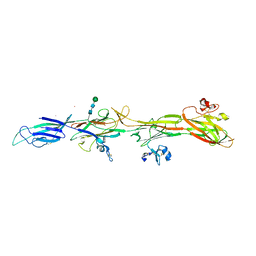 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | Descriptor: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | Authors: | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
7MJU
 
 | |
2LDC
 
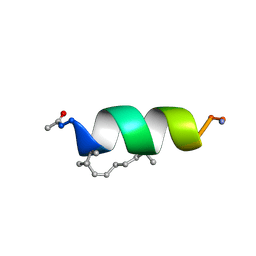 | | Solution structure of the estrogen receptor-binding stapled peptide SP1 (Ac-HXILHXLLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP1 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2LDD
 
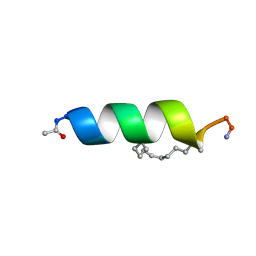 | | Solution structure of the estrogen receptor-binding stapled peptide SP6 (Ac-EKHKILXRLLXDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP6 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2LDA
 
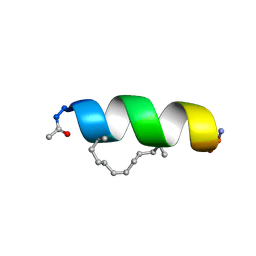 | | Solution structure of the estrogen receptor-binding stapled peptide SP2 (Ac-HKXLHQXLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP2 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
7OI0
 
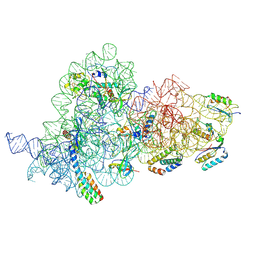 | | E.coli delta rbfA pre-30S ribosomal subunit class D | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Maksimova, E, Korepanov, A, Baymukhametov, T, Kravchenko, O, Stolboushkina, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center.
Int J Mol Sci, 22, 2021
|
|
4UAL
 
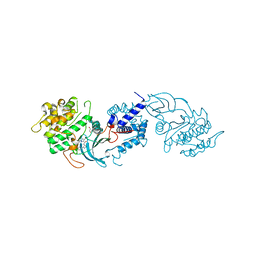 | | MRCK beta in complex with BDP00005290 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-1-(piperidin-4-yl)-N-[3-(pyridin-2-yl)-1H-pyrazol-4-yl]-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2014-08-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A novel small-molecule MRCK inhibitor blocks cancer cell invasion.
Cell Commun. Signal, 12, 2014
|
|
4UAK
 
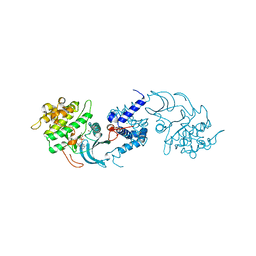 | | MRCK beta in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2014-08-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A novel small-molecule MRCK inhibitor blocks cancer cell invasion.
Cell Commun. Signal, 12, 2014
|
|
8BQU
 
 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament | | Descriptor: | Choriogenin H, Zona pellucida sperm-binding protein 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bokhove, M, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2022-11-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
3H3F
 
 | | Rabbit muscle L-lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Bujacz, A, Bujacz, G, Swiderek, K, Paneth, P. | | Deposit date: | 2009-04-16 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modeling of isotope effects on binding oxamate to lactic dehydrogenase
J.Phys.Chem.B, 113, 2009
|
|
6ODL
 
 | | Crystal structure of GluN2A agonist binding domain with 4-butyl-(S)-CCG-IV | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]-1-butylcyclopropane-1-carboxylic acid, Glutamate receptor ionotropic, NMDA 2A,Glutamate receptor ionotropic, ... | | Authors: | Mou, T.C, Clausen, R.P, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stereoselective synthesis of novel 2'-(S)-CCG-IV analogues as potent NMDA receptor agonists.
Eur.J.Med.Chem., 212, 2021
|
|
