7TL0
 
 | | Cryo-EM structure of hMPV preF bound by Fabs MPE8 and SAN32-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Rush, S.A, Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Characterization of prefusion-F-specific antibodies elicited by natural infection with human metapneumovirus.
Cell Rep, 40, 2022
|
|
7UR4
 
 | |
6VKF
 
 | | CCHFV GP38 (IbAr10200) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GP38 | | Authors: | Mishra, A.K, McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.524 Å) | | Cite: | Structure and Characterization of Crimean-Congo Hemorrhagic Fever Virus GP38.
J.Virol., 94, 2020
|
|
8TCO
 
 | | HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CS2it1p2_F7K Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2023-07-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Single-cell analysis of memory B cells from top neutralizers reveals multiple sites of vulnerability within HCMV Trimer and Pentamer.
Immunity, 56, 2023
|
|
7UOP
 
 | |
7UP9
 
 | | Prefusion-stabilized Nipah virus fusion protein complexed with Fab 2D3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2D3 heavy chain, ... | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for antibody recognition of vulnerable epitopes on Nipah virus F protein.
Nat Commun, 14, 2023
|
|
7UPD
 
 | |
7UPA
 
 | |
7UPB
 
 | |
7UPK
 
 | |
6WAQ
 
 | |
6WAR
 
 | |
6VKM
 
 | | Crystal Structure of Stabilized GP from Makona Variant of Ebola Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Virion spike glycoprotein | | Authors: | Gilman, M.S.A, Rutten, L, Langedijk, J.P.M, McLellan, J.S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Based Design of Prefusion-Stabilized Filovirus Glycoprotein Trimers.
Cell Rep, 30, 2020
|
|
6VSB
 
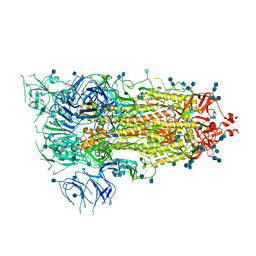 | | Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrapp, D, Wang, N, Corbett, K.S, Goldsmith, J.A, Hsieh, C, Abiona, O, Graham, B.S, McLellan, J.S. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-26 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation.
Science, 367, 2020
|
|
5WB0
 
 | |
5W23
 
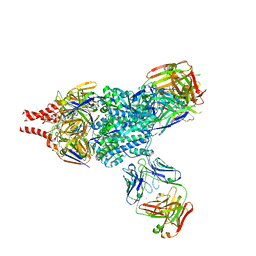 | | Crystal Structure of RSV F in complex with 5C4 Fab | | Descriptor: | 5C4 Fab heavy chain, 5C4 Fab light chain, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S. | | Deposit date: | 2017-06-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein.
Nat Commun, 8, 2017
|
|
5VYH
 
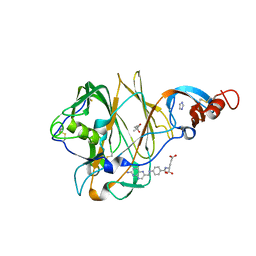 | | Crystal Structure of MERS-CoV S1 N-terminal Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Wang, N, Wrapp, D, Pallesen, J, Ward, A.B, McLellan, J.S. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VZR
 
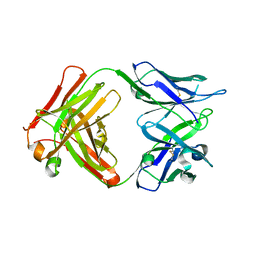 | |
6DC4
 
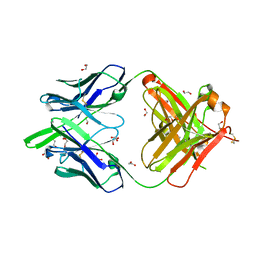 | | RSV-neutralizing human antibody AM22 | | Descriptor: | 1,2-ETHANEDIOL, Fab AM22 Heavy Chain, Fab AM22 Light Chain | | Authors: | Jones, H.G, McLellan, J.S. | | Deposit date: | 2018-05-04 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternative conformations of a major antigenic site on RSV F.
Plos Pathog., 15, 2019
|
|
6DC3
 
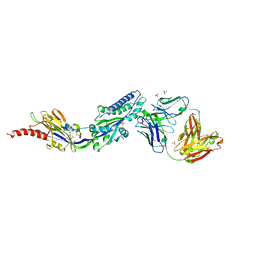 | | RSV prefusion F bound to RSD5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab RSD5-Germline Heavy Chain, Fab RSD5-Germline Light Chain, ... | | Authors: | Battles, M.B, McLellan, J.S, Jones, H.J. | | Deposit date: | 2018-05-04 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Alternative conformations of a major antigenic site on RSV F.
Plos Pathog., 15, 2019
|
|
6DC5
 
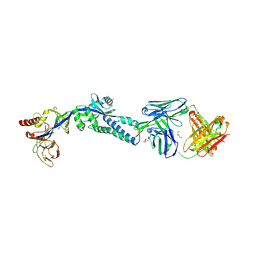 | | RSV prefusion F in complex with AM22 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jones, H.G, McLellan, J.S. | | Deposit date: | 2018-05-04 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Alternative conformations of a major antigenic site on RSV F.
Plos Pathog., 15, 2019
|
|
6EAH
 
 | |
6EAL
 
 | |
6EAI
 
 | |
6EAM
 
 | |
