5CIX
 
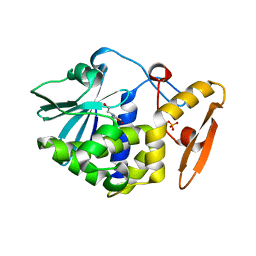 | | Structure of the complex of type 1 Ribosome inactivating protein with triethanolamine at 1.88 Angstrom resolution | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Singh, P.K, Pandey, S, Tyagi, T.K, Singh, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of the complex of type 1 Ribosome inactivating protein with triethanolamine at 1.88 Angstrom resolution.
To Be Published
|
|
5CRY
 
 | | Structure of iron-saturated C-lobe of bovine lactoferrin at pH 6.8 indicates the softening of iron coordination | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Singh, A, Rastogi, N, Singh, P.K, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of iron-saturated C-lobe of bovine lactoferrin at pH 7.0 indicates the softening of iron coordination
To Be Published
|
|
5CSO
 
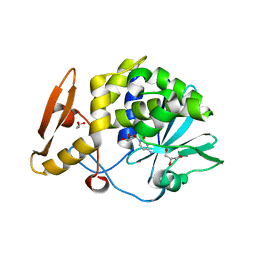 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleoside, cytidine at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
5CST
 
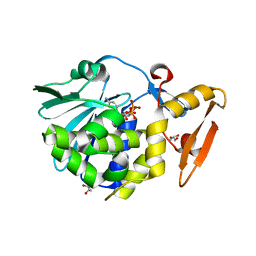 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine diphosphate at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
2PI6
 
 | | Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chitinase-3-like protein 1, ETHANOL, ... | | Authors: | Sharma, P, Singh, N, Sharma, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2007-04-13 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Tryptophan as a three-way switch in regulating the function of the secretory signalling glycoprotein (SPS-40) from mammary glands: structure of SPS-40 complexed with 2-methylpentane-2,4-diol at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2O92
 
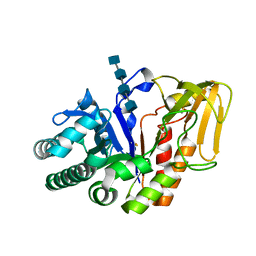 | | Crystal structure of a signalling protein (SPG-40) complex with tetrasaccharide at 3.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sharma, P, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-12-13 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a signalling protein (SPG-40) complex with tetrasaccharide at 3.0A resolution
To be Published
|
|
2OLH
 
 | | Crystal structure of a signalling protein (SPG-40) complex with cellobiose at 2.78 A resolution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Sharma, P, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-01-19 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a signalling protein (SPG-40) complex with cellobiose at 2.78 A resolution
To be Published
|
|
2R2K
 
 | | Crystal structure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein | | Authors: | Sharma, P, Jain, R, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-08-26 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution
To be Published
|
|
2R90
 
 | | Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution | | Descriptor: | Peptidoglycan recognition protein | | Authors: | Sharma, P, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-09-12 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution
To be Published
|
|
3I6N
 
 | | Mode of Binding of the Tuberculosis Prodrug Isoniazid to Peroxidases: Crystal Structure of Bovine Lactoperoxidase with Isoniazid at 2.7 Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIAZENYLCARBONYL)PYRIDINE, CALCIUM ION, ... | | Authors: | Singh, A.K, Kumar, R.P, Pandey, N, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2009-07-07 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution.
J.Biol.Chem., 285, 2010
|
|
3HU7
 
 | | Structural characterization and binding studies of a plant pathogenesis related protein heamanthin from haemanthus multiflorus reveal its dual inhibitory effects against xylanase and alpha-amylase | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-06-13 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
3IB1
 
 | | Structural basis of the prevention of NSAID-induced damage of the gastrointestinal tract by C-terminal half (C-lobe) of bovine colostrum protein lactoferrin: Binding and structural studies of C-lobe complex with indomethacin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for the prevention of nonsteroidal antiinflammatory drug-induced gastrointestinal tract damage by the C-lobe of bovine colostrum lactoferrin
Biophys.J., 97, 2009
|
|
3GCI
 
 | | Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution | | Descriptor: | CALCIUM ION, Heptapeptide from Amyloid beta A4 protein, Phospholipase A2 isoform 3 | | Authors: | Mirza, Z, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution
To be Published
|
|
3GC1
 
 | | Crystal structure of bovine lactoperoxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-21 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution.
J.Biol.Chem., 285, 2010
|
|
3GCL
 
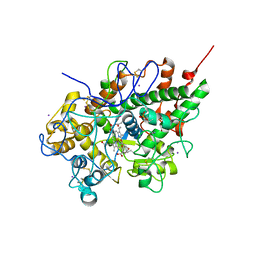 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding modes of aromatic ligands to mammalian heme peroxidases with associated functional implications: crystal structures of lactoperoxidase complexes with acetylsalicylic acid, salicylhydroxamic acid, and benzylhydroxamic acid
J.Biol.Chem., 284, 2009
|
|
3IB0
 
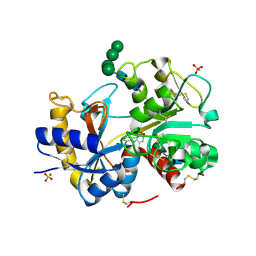 | | Structural basis of the prevention of NSAID-induced damage of the gastrointestinal tract by C-terminal half (C-lobe) of bovine colostrum protein lactoferrin: Binding and structural studies of C-lobe complex with diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structural basis for the prevention of nonsteroidal antiinflammatory drug-induced gastrointestinal tract damage by the C-lobe of bovine colostrum lactoferrin
Biophys.J., 97, 2009
|
|
3IAZ
 
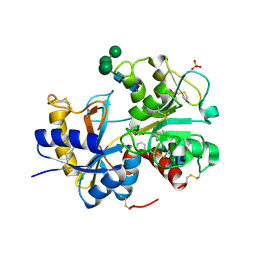 | | Structural basis of the prevention of NSAID-induced damage of the gastrointestinal tract by C-terminal half (C-lobe) of bovine colostrum protein lactoferrin: Binding and structural studies of the C-lobe complex with aspirin | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for the prevention of nonsteroidal antiinflammatory drug-induced gastrointestinal tract damage by the C-lobe of bovine colostrum lactoferrin
Biophys.J., 97, 2009
|
|
3G8F
 
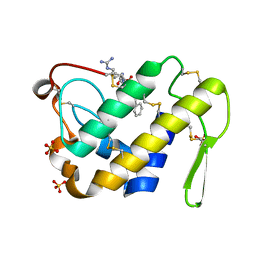 | | Crystal structure of the complex formed between a group II phospholipase A2 and designed peptide inhibitor carbobenzoxy-dehydro-val-ala-arg-ser at 1.2 A resolution | | Descriptor: | PHQ VAL ALA ARG SER peptide, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Kaur, P, Prem Kumar, R, Somvanshi, R.K, Perbandt, M, Betzel, C, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the Complex Formed between a Group II Phospholipase A2 and Designed Peptide Inhibitor Carbobenzoxy-Dehydro-Val-Ala-Arg-Ser at 1.2 A Resolution
To be Published
|
|
3GCJ
 
 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid
To be Published
|
|
3GCK
 
 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid
To be Published
|
|
3H1X
 
 | | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site | | Descriptor: | INDOMETHACIN, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Prem Kumar, R, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2009-04-14 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simultaneous inhibition of anti-coagulation and inflammation: crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand-binding site
J.Mol.Recognit., 22, 2009
|
|
3NG4
 
 | | Ternary complex of peptidoglycan recognition protein (PGRP-S) with Maltose and N-Acetylglucosamine at 1.7 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3JQL
 
 | | Crystal Structure of the Complex Formed Between Phospholipase A2 and a Hexapeptide Fragment of Amyloid Beta Peptide, Lys-Leu-Val-Phe-Phe-Ala at 1.2 A Resolution | | Descriptor: | Acidic phospholipase A2 3 (Fragment), Amyloid Beta Peptide, CALCIUM ION | | Authors: | Mirza, Z, Vikram, G, Singh, N, Sinha, M, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2009-09-07 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of the Complex Formed Between Phospholipase A2 and a Hexapeptide Fragment of Amyloid Beta Peptide, Lys-Leu-Val-Phe-Phe-Ala at 1.2 A Resolution
To be Published
|
|
3JTI
 
 | | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 3, octapeptide from Amyloid beta A4 protein | | Authors: | Pandey, N, Mirza, Z, Vikram, G, Singh, N, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-09-12 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution
To be Published
|
|
3IB2
 
 | | structure of the complex of C-terminal half (C-lobe) of bovine lactoferrin with alpha-methyl-4-(2-methylpropyl) benzene acetic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Vikram, G, Kumar, R.P, Sinha, M, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2009-07-15 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The structural basis for the prevention of nonsteroidal antiinflammatory drug-induced gastrointestinal tract damage by the C-lobe of bovine colostrum lactoferrin.
Biophys.J., 97, 2009
|
|
