2MSY
 
 | | Solution structure of Hox homeodomain | | Descriptor: | Homeobox protein Hox-C9 | | Authors: | Kim, H, Park, S, Han, J, Lee, B. | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the interaction between the Hox and HMGB1 and understanding of the HMGB1-enhancing effect of Hox-DNA binding.
Biochim.Biophys.Acta, 1854, 2015
|
|
3OUH
 
 | | PHD2-R127 with JNJ41536014 | | Descriptor: | 1-(5-chloro-6-fluoro-1H-benzimidazol-2-yl)-1H-pyrazole-4-carboxylic acid, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Kim, H, Clark, R. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-01 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues.
ACS Med Chem Lett, 1, 2010
|
|
8HEJ
 
 | | Crystal structure of Transthyretin in complex with a covalent inhibitor trans-styrylpyrazole | | Descriptor: | 2,4,6-trifluorobenzaldehyde, 2,6-dibromo-4-[(E)-2-(3,5-dimethyl-1H-pyrazol-4-yl)ethenyl]phenol, Transthyretin | | Authors: | Kim, H, Choi, S, Lee, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of Transthyretin in complex with a covalent inhibitor trans-styrylpyrazole
To Be Published
|
|
5B4O
 
 | | Crystal structure of Macrophage Migration Inhibitory Factor in complex with BTZO-14 | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-1,3-benzothiazin-4-one, Macrophage migration inhibitory factor, ... | | Authors: | Oki, H, Igaki, S, Moriya, Y, Hayano, Y, Habuka, N. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression
Chem. Biol., 17, 2010
|
|
7P80
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compressed state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7P81
 
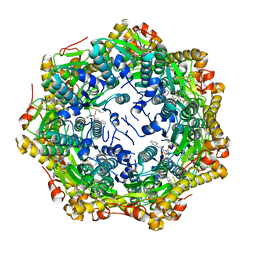 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
5XTG
 
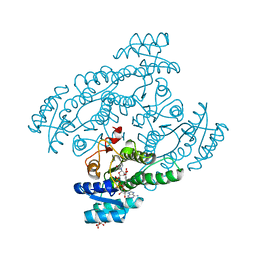 | | Crystal structure of the cis-dihydrodiol naphthalene dehydrogenase NahB from Pseudomonas sp. MC1 in the presence of NAD+ and 2,3-dihydroxybiphenyl | | Descriptor: | 2,3-dihydroxy-2,3-dihydrophenylpropionate dehydrogenase, BIPHENYL-2,3-DIOL, CITRIC ACID, ... | | Authors: | Park, A.K, Kim, H.-W. | | Deposit date: | 2017-06-19 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.318 Å) | | Cite: | Crystal structure of cis-dihydrodiol naphthalene dehydrogenase (NahB) from Pseudomonas sp. MC1: Insights into the early binding process of the substrate
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5XTF
 
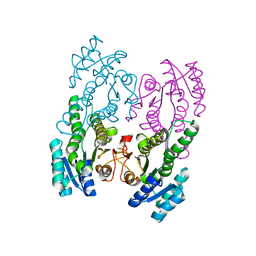 | |
2KDC
 
 | | NMR Solution Structure of E. coli diacylglycerol kinase (DAGK) in DPC micelles | | Descriptor: | Diacylglycerol kinase | | Authors: | Van Horn, W.D, Kim, H, Ellis, C.D, Hadziselimovic, A, Sulistijo, E.S, Karra, M.D, Tian, C, Sonnichsen, F.D, Sanders, C.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase
Science, 324, 2009
|
|
6PLN
 
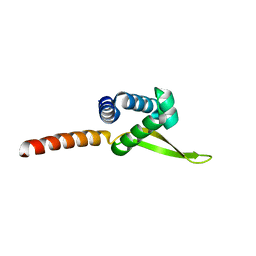 | |
6IJR
 
 | | Human PPARgamma ligand binding domain complexed with SB1495 | | Descriptor: | 16 mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1S,2R)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6IJS
 
 | | Human PPARgamma ligand binding domain complexed with SB1494 | | Descriptor: | 16-mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1R,2S)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
8HFB
 
 | | Evolved variant of quercetin 2,4-dioxygenase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Eom, H, Song, W.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Underlying Role of Hydrophobic Environments in Tuning Metal Elements for Efficient Enzyme Catalysis.
J.Am.Chem.Soc., 145, 2023
|
|
5XB1
 
 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Ahn, B.J, Jeong, H, Kim, H, Hyun, J, Jung, Y. | | Deposit date: | 2017-03-15 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4UIP
 
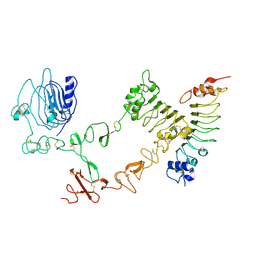 | | The complex structure of extracellular domain of EGFR with Repebody (rAC1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Kang, Y.J, Cha, Y.J, Cho, H.S, Lee, J.J, Kim, H.S. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Enzymatic Prenylation and Oxime Ligation for the Synthesis of Stable and Homogeneous Protein-Drug Conjugates for Targeted Therapy.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1A77
 
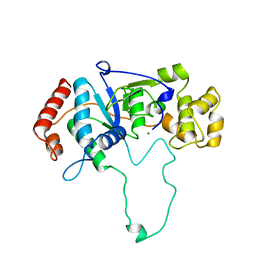 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MAGNESIUM ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
6RMH
 
 | | The Rigid-body refined model of the normal Huntingtin. | | Descriptor: | Huntingtin | | Authors: | Jung, T, Tamo, G, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2019-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
1A76
 
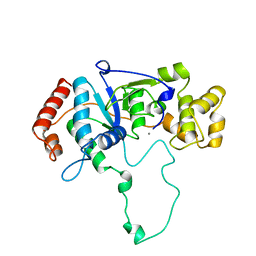 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MANGANESE (II) ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
8IVU
 
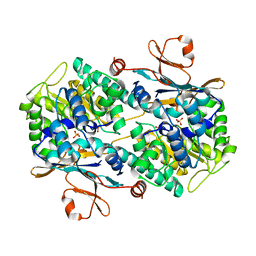 | | Crystal Structure of Human NAMPT in complex with A4276 | | Descriptor: | N-[[4-(6-methyl-1,3-benzoxazol-2-yl)phenyl]methyl]pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Kang, B.G, Cha, S.S. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09000921 Å) | | Cite: | Discovery of a novel NAMPT inhibitor that selectively targets NAPRT-deficient EMT-subtype cancer cells and alleviates chemotherapy-induced peripheral neuropathy.
Theranostics, 13, 2023
|
|
5G54
 
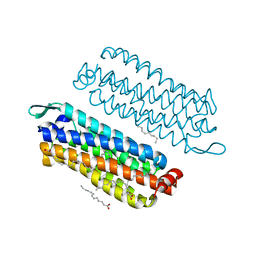 | | The crystal structure of light-driven chloride pump ClR at pH 4.5 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
8J26
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-4, AM047-6, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
8J1Q
 
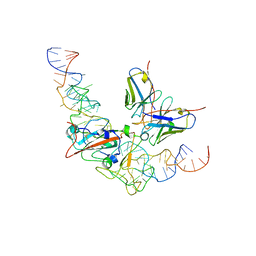 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-0, AM047-0, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
5G28
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2D
 
 | | The crystal structure of light-driven chloride pump ClR (T102N) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMP RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2C
 
 | | The crystal structure of light-driven chloride pump ClR (T102D) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
