3AEX
 
 | |
3AEY
 
 | |
2CWV
 
 | | Product schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWU
 
 | | Substrate schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWT
 
 | | Catalytic base deletion in copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
1WII
 
 | | Solution structure of RSGI RUH-025, a DUF701 domain from mouse cDNA | | Descriptor: | Hypothetical UPF0222 protein MGC4549, ZINC ION | | Authors: | Abe, T, Hirota, H, Hayashi, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-025, a DUF701 domain from mouse cDNA
To be Published
|
|
2E2U
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with 4-hydroxybenzylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
2D1W
 
 | | Substrate Schiff-Base intermediate with tyramine in copper amine oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Kuroda, S, Nakamoto, T, Taki, M, Yamamoto, Y, Hayashi, H, Tanizawa, K. | | Deposit date: | 2005-09-01 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Quantum mechanical hydrogen tunneling in bacterial copper amine oxidase reaction
Biochem.Biophys.Res.Commun., 342, 2006
|
|
2E2V
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with benzylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Kuroda, S, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
2E2T
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with phenylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Kuroda, S, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
7QRO
 
 | |
1AY4
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITHOUT SUBSTRATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY5
 
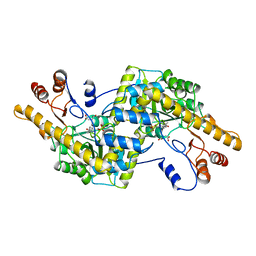 | | AROMATIC AMINO ACID AMINOTRANSFERASE COMPLEX WITH MALEATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY8
 
 | |
7JKV
 
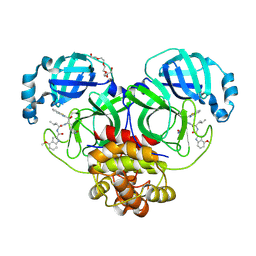 | | Crystal Structure of SARS-CoV-2 main protease in complex with an inhibitor GRL-2420 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, PENTAETHYLENE GLYCOL | | Authors: | Bulut, H, Hattori, S.I, Das, D, Murayama, K, Mitsuya, H. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication.
Nat Commun, 12, 2021
|
|
5F9E
 
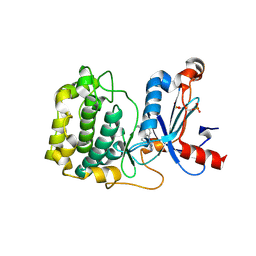 | | Structure of Protein Kinase C theta with compound 10: 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one | | Descriptor: | 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one, Protein kinase C theta type | | Authors: | Klein, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of 1,7-disubstituted-2,2-dimethyl-2,3-dihydroquinazolin-4(1H)-ones as potent and selective PKC theta inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
6E7J
 
 | | HIV-1 wild type protease with GRL-042-17A, 3-phenylhexahydro-2h-cyclopenta[d]oxazol-2-one with a bicyclic oxazolidinone scaffold as the P2 ligand | | Descriptor: | (3aS,5R,6aR)-2-oxo-3-phenylhexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
6E9A
 
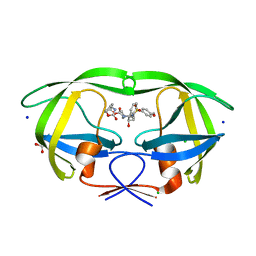 | | HIV-1 WILD TYPE PROTEASE WITH GRL-034-17A, (3aS, 5R, 6aR)-2-OXOHEXAHYD CYCLOPENTA[D]-5-OXAZOLYL URETHANE WITH A BICYCLIC OXAZOLIDINONE SCAFF AS THE P2 LIGAND | | Descriptor: | (3aS,5R,6aR)-2-oxohexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
5ULT
 
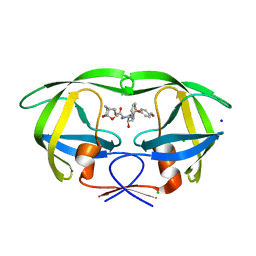 | | HIV-1 wild Type protease with GRL-100-13A (a Crown-like Oxotricyclic Core as the P2-Ligand with the sulfonamide isostere as the P2' group) | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and Development of Highly Potent HIV-1 Protease Inhibitors with a Crown-Like Oxotricyclic Core as the P2-Ligand To Combat Multidrug-Resistant HIV Variants.
J. Med. Chem., 60, 2017
|
|
1G4X
 
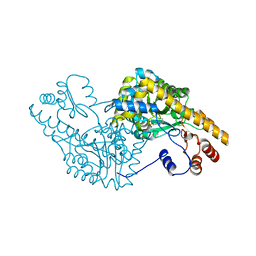 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
5XDF
 
 | |
6DUW
 
 | |
6DV1
 
 | |
6DUY
 
 | |
5HXI
 
 | | 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5HN bound | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Mikami, B. | | Deposit date: | 2016-01-30 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of the Tyr270 residue in 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti
J. Biosci. Bioeng., 123, 2017
|
|
