1G4R
 
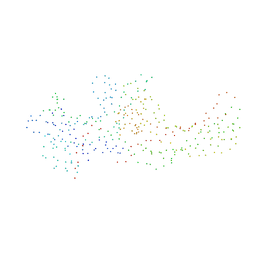 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN 1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
1G4M
 
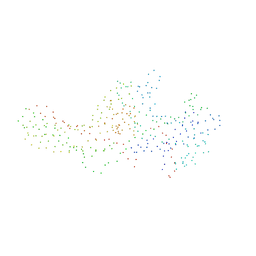 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
6INW
 
 | | A Pericyclic Reaction enzyme | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Feng, Y, Chang, M, Wang, H, Liu, Z, Zhou, Y. | | Deposit date: | 2018-10-28 | | Release date: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of the multifunctional SAM-dependent enzyme LepI provides insights into its catalytic mechanism.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
8HDK
 
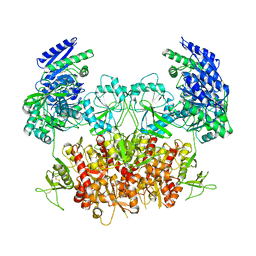 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (minor class in symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7VRD
 
 | |
7V67
 
 | |
6XJ1
 
 | |
6EHF
 
 | |
6EHE
 
 | |
6EHB
 
 | |
6EHC
 
 | |
6EHD
 
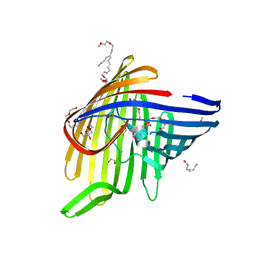 | |
7A4A
 
 | |
8SUV
 
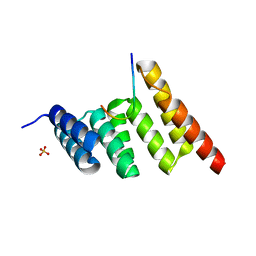 | | CHIP-TPR in complex with the C-terminus of CHIC2 | | Descriptor: | Cysteine-rich hydrophobic domain-containing protein 2, E3 ubiquitin-protein ligase CHIP, SULFATE ION | | Authors: | Cupo, A.R, McDermott, L.E, DeSilva, A.R, Callahan, M, Nix, J.C, Gestwicki, J.E, Page, R.C. | | Deposit date: | 2023-05-13 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Interaction with the membrane-anchored protein CHIC2 constrains the ubiquitin ligase activity of CHIP
Biorxiv, 2023
|
|
5BT1
 
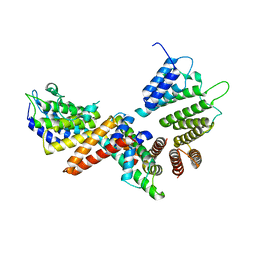 | | histone chaperone Hif1 playing with histone H2A-H2B dimer | | Descriptor: | HAT1-interacting factor 1, Histone H2A.1, Histone H2B.1 | | Authors: | Liu, H, Zhang, M, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2015-06-02 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer
Structure, 24, 2016
|
|
6JQF
 
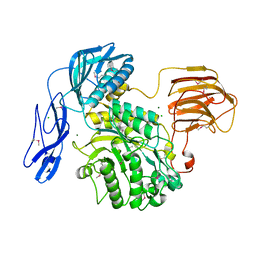 | |
8PF9
 
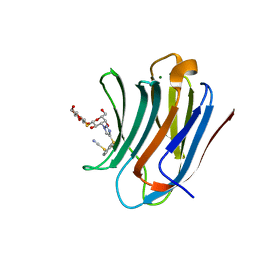 | | Galectin-3C in complex with a triazolesulfane derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[oxidanyl(phenyl)-$l^{3}-sulfanyl]-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8PBF
 
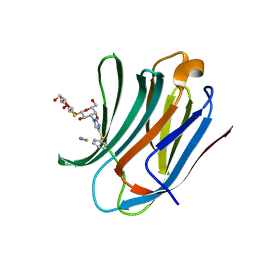 | | Galectin-3C in complex with a triazolesulfane derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-(4-phenylsulfanyl-1,2,3-triazol-1-yl)oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Mahanti, M. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8PFF
 
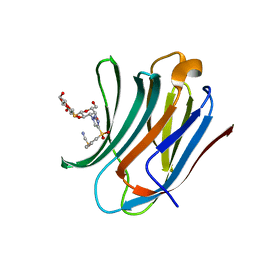 | | Galectin-3C in complex with a triazolesulfone derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-(phenylsulfonyl)-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
1TJ2
 
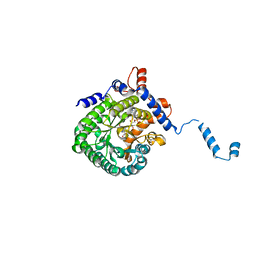 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with acetate | | Descriptor: | ACETATE ION, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
7X1N
 
 | | Crystal structure of MEF2D-MRE complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte enhancer factor 2D/deleted in azoospermia associated protein 1 fusion protein | | Authors: | Zhang, H, Zhang, M, Wang, Q.Q, Chen, Z, Chen, S.J, Meng, G. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.315 Å) | | Cite: | Functional, structural, and molecular characterizations of the leukemogenic driver MEF2D-HNRNPUL1 fusion.
Blood, 140, 2022
|
|
6JAO
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in complex with palatinose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6JPF
 
 | |
6JAQ
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6JAI
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein D118A in complex with maltose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
