1TGJ
 
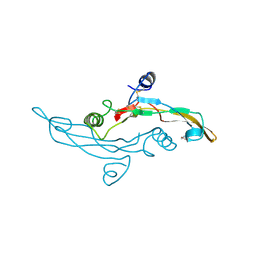 | | HUMAN TRANSFORMING GROWTH FACTOR-BETA 3, CRYSTALLIZED FROM DIOXANE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, TRANSFORMING GROWTH FACTOR-BETA 3 | | Authors: | Mittl, P.R.E, Priestle, J.P, Gruetter, M.G. | | Deposit date: | 1996-07-09 | | Release date: | 1997-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of TGF-beta 3 and comparison to TGF-beta 2: implications for receptor binding.
Protein Sci., 5, 1996
|
|
1L10
 
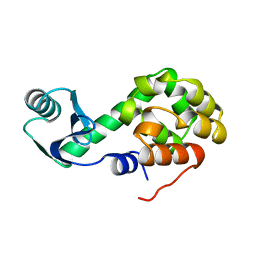 | |
1L01
 
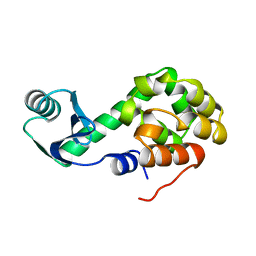 | |
1L02
 
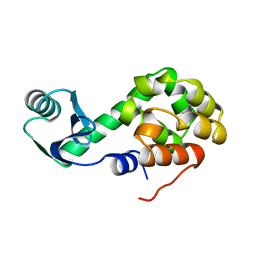 | |
1L09
 
 | |
1L06
 
 | |
1L15
 
 | |
1L13
 
 | |
1L12
 
 | |
1L16
 
 | |
1L03
 
 | | CONTRIBUTIONS OF HYDROGEN BONDS OF THR 157 TO THE THERMODYNAMIC STABILITY OF PHAGE T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Dao-Pin, S, Wilson, K, Alber, T, Matthews, B.W. | | Deposit date: | 1988-02-05 | | Release date: | 1988-04-16 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme.
Nature, 330, 1987
|
|
1L07
 
 | |
1L08
 
 | |
1L04
 
 | |
1L11
 
 | | CONTRIBUTIONS OF HYDROGEN BONDS OF THR 157 TO THE THERMODYNAMIC STABILITY OF PHAGE T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Dao-Pin, S, Wilson, K, Alber, T, Matthews, B.W. | | Deposit date: | 1988-02-05 | | Release date: | 1988-04-16 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme.
Nature, 330, 1987
|
|
1L14
 
 | |
1L05
 
 | |
154L
 
 | |
1C94
 
 | | REVERSING THE SEQUENCE OF THE GCN4 LEUCINE ZIPPER DOES NOT AFFECT ITS FOLD. | | Descriptor: | RETRO-GCN4 LEUCINE ZIPPER | | Authors: | Mittl, P.R.E, Deillon, C.A, Sargent, D, Liu, N, Klauser, S, Thomas, R.M, Gutte, B, Gruetter, M.G. | | Deposit date: | 1999-07-30 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
153L
 
 | |
1CP3
 
 | |
3EO0
 
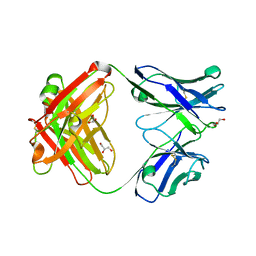 | |
2C2M
 
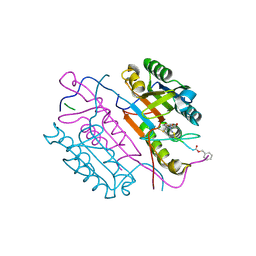 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-[4-(BENZYLOXY)-4-OXOBUTANOYL]-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14 -PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
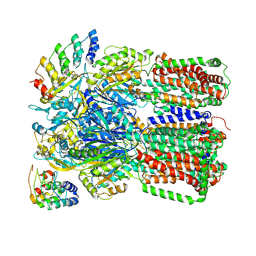
3NOG
 
 | |
2C2O
 
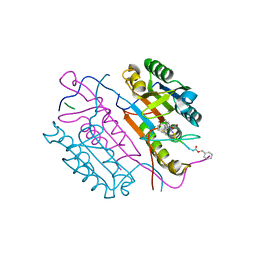 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-{4-[BENZYL(METHYL) AMINO]-4-OXOBUTANOYL}-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
