2NYR
 
 | | Crystal Structure of Human Sirtuin Homolog 5 in Complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, NAD-dependent deacetylase sirtuin-5, ZINC ION | | Authors: | Min, J.R, Antoshenko, T, Allali-Hassani, A, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin.
Structure, 15, 2007
|
|
4YO1
 
 | |
4YNN
 
 | | Structure of Legionella pneumophila DegQ (S190A variant) | | Descriptor: | Hepta-peptide, Hexa-peptide, Octapeptide, ... | | Authors: | Wrase, R, Hilgenfeld, R, Hansen, G. | | Deposit date: | 2015-03-10 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of DegQ from Legionella pneumophila Define Distinct ON and OFF States.
J.Mol.Biol., 427, 2015
|
|
6XNN
 
 | | Crystal Structure of Mouse STING CTD complex with SR-717. | | Descriptor: | 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, Stimulator of interferon genes protein | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
6XNP
 
 | | Crystal Structure of Human STING CTD complex with SR-717 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, GLYCEROL, ... | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
6Q0C
 
 | | MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with undamaged dG | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, CALCIUM ION, ... | | Authors: | O'Shea Murray, V.L, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2019-08-01 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Acs Chem.Biol., 15, 2020
|
|
4K7P
 
 | |
7LMO
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 34 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-7-(1H-imidazole-5-carbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione] | | Descriptor: | (5~{R})-3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-7-(1~{H}-imidazol-4-ylcarbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7LMN
 
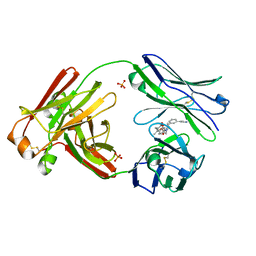 | |
7LMR
 
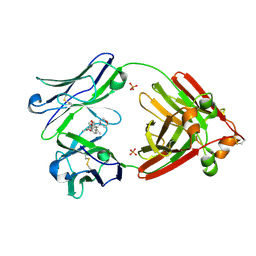 | |
7LMP
 
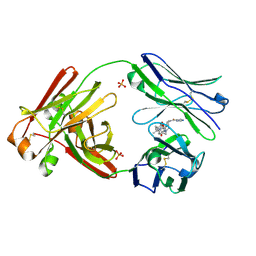 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 36 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-8-(1H-imidazole-4-carbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione] | | Descriptor: | 3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-8-(1~{H}-imidazol-5-ylcarbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7LMQ
 
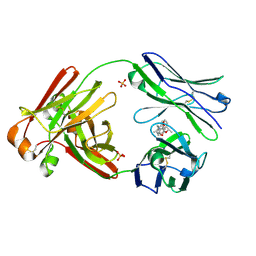 | |
6SKE
 
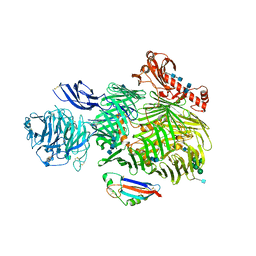 | | Teneurin 2 in complex with Latrophilin 2 Lec domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor L2, ... | | Authors: | Shahin, M, Jackson, V.A, Carrasquero, M, Lowe, E, Seiradake, E. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural Basis of Teneurin-Latrophilin Interaction in Repulsive Guidance of Migrating Neurons.
Cell, 180, 2020
|
|
7RTP
 
 | |
6S3Q
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3)
TO BE PUBLISHED
|
|
8QZ7
 
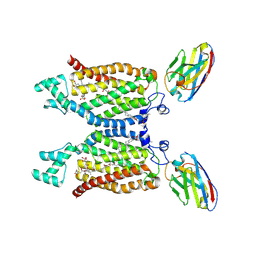 | | Structure of human ceramide synthase 6 (CerS6) in complex with N-palmitoyl fumonisin B1 | | Descriptor: | (2~{R})-2-[2-[(5~{R},6~{R},7~{S},9~{S},11~{R},16~{R},18~{S},19~{S})-6-[(3~{R})-3-carboxy-5-oxidanyl-5-oxidanylidene-pentanoyl]oxy-19-(hexadecanoylamino)-5,9-dimethyl-11,16,18-tris(oxidanyl)icosan-7-yl]oxy-2-oxidanylidene-ethyl]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase
Nature Structural & Molecular Biology, 2024
|
|
8QZ6
 
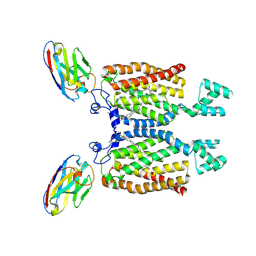 | | Structure of human ceramide synthase 6 (CerS6) bound to C16:0 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Ceramide synthase 6, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase
Nature Structural & Molecular Biology, 2024
|
|
9EOT
 
 | | Structure of human ceramide synthase 6 (CerS6) bound to C16:0 (nanobody Nb02) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Ceramide synthase 6, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase
Nature Structural & Molecular Biology, 2024
|
|
6U7T
 
 | | MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G) | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | Authors: | O'Shea Murray, V.L, Cao, S, Horvath, M.P, David, S.S. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Acs Chem.Biol., 15, 2020
|
|
1SES
 
 | |
7O45
 
 | | Crystal structure of ADD domain of the human DNMT3B methyltransferase | | Descriptor: | BROMIDE ION, Isoform 6 of DNA (cytosine-5)-methyltransferase 3B, ZINC ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-04-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the DNMT3B ADD domain suggests the absence of a DNMT3A-like autoinhibitory mechanism.
Biochem.Biophys.Res.Commun., 619, 2022
|
|
8QX6
 
 | | Novel laminarin-binding CBM X584 | | Descriptor: | PKD domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Zuehlke, M.K, Jeudy, A, Czjzek, M. | | Deposit date: | 2023-10-22 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ.Microbiol., 26, 2024
|
|
1SET
 
 | |
8DD5
 
 | | Crystal structure of KAT6A in complex with inhibitor CTx-648 (PF-9363) | | Descriptor: | 2,6-dimethoxy-N-{4-methoxy-6-[(1H-pyrazol-1-yl)methyl]-1,2-benzoxazol-3-yl}benzene-1-sulfonamide, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Greasley, S.E, Johnson, E, Brodsky, O. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Targeting KAT6A/KAT6B dependencies in breast cancer with a novel selective, orally bioavailable KAT6 inhibitor, CTx-648/PF-9363
To Be Published
|
|
7O6T
 
 | | Crystal structure of the polymerising VEL domain of VIN3 (R556D I575D mutant) | | Descriptor: | MAGNESIUM ION, Protein VERNALIZATION INSENSITIVE 3 | | Authors: | Fiedler, M, Franco-Echevarria, E, Dean, C, Bienz, M. | | Deposit date: | 2021-04-12 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Head-to-tail polymerization by VEL proteins underpins cold-induced Polycomb silencing in flowering control.
Cell Rep, 41, 2022
|
|
